Search Count: 1,246
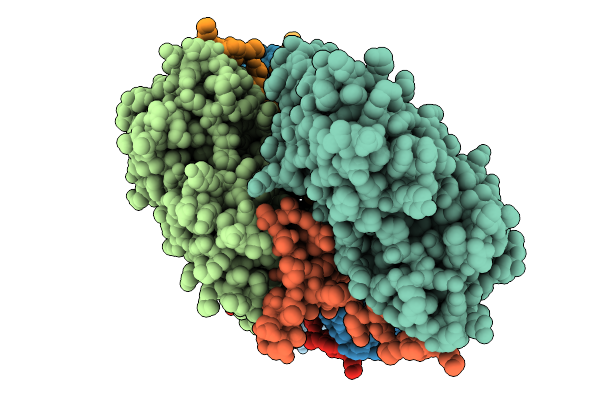 |
Atomic Structure Of Vibrio Effector Fragment Vopv Bound To Beta-Cytoplasmic/Gamma1-Cytoplasmic F-Actin
Organism: Homo sapiens, Vibrio parahaemolyticus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: STRUCTURAL PROTEIN Ligands: MG, ANP |
 |
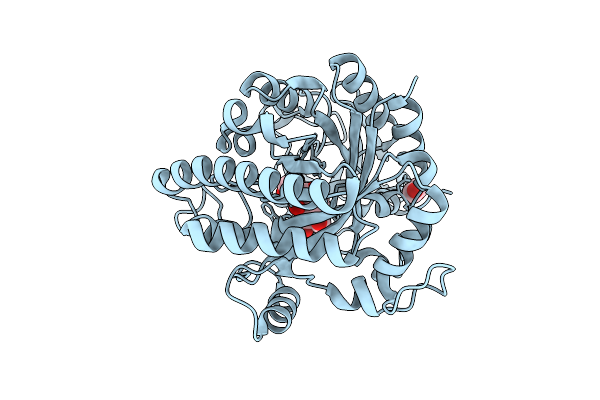
Organism: Vibrio parahaemolyticus
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: HYDROLASE |
 |
Organism: Vibrio parahaemolyticus
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: EDO, B3P |
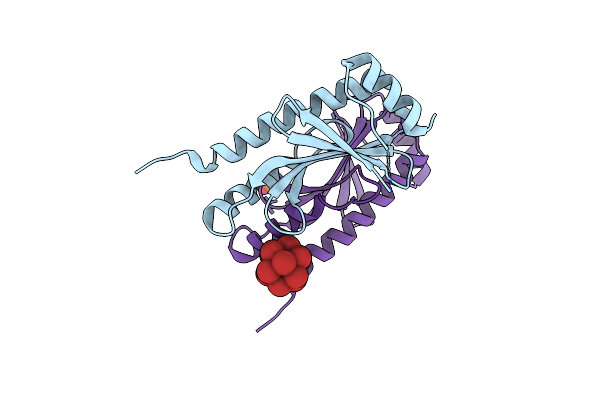 |
Crystal Structure Of The Type Iii Secretion Chaperone Veca From Vibrio Parahaemolyticus
Organism: Vibrio parahaemolyticus
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: CHAPERONE Ligands: PO4, TBR |
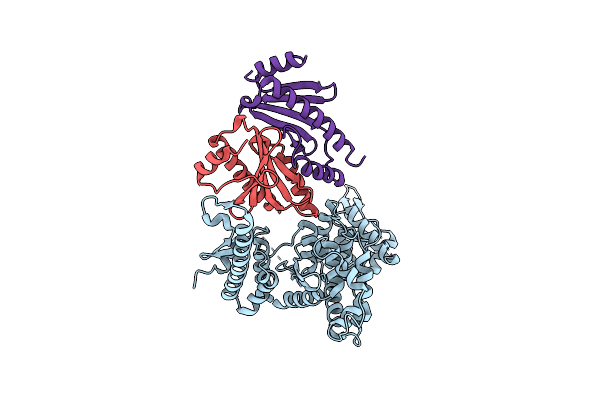 |
Crystal Structure Of The Virulence Effector Vepa In Complex With Its Secretion Chaperone Veca
Organism: Vibrio parahaemolyticus
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: TOXIN |
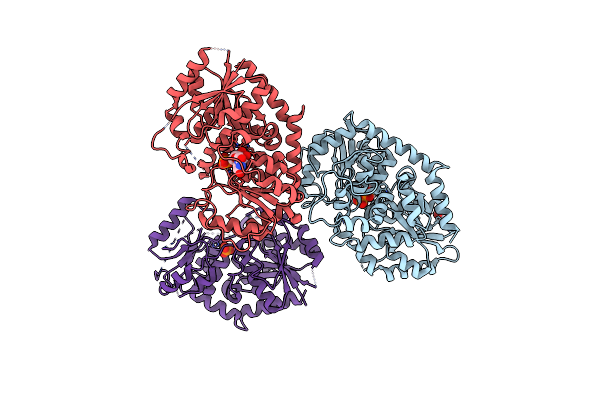 |
Organism: Stevia rebaudiana
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: TRANSFERASE Ligands: UPG, IPA, PO4 |
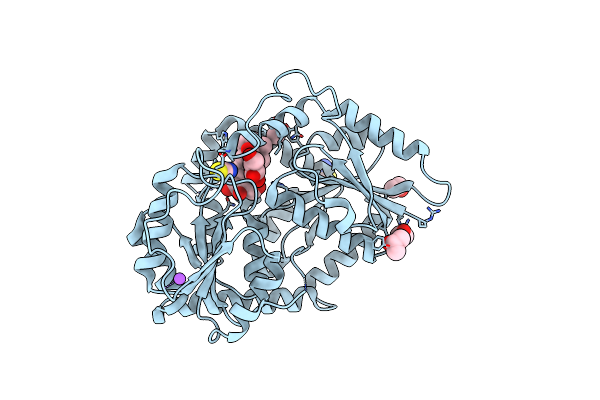 |
Organism: Stevia rebaudiana
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: TRANSFERASE |
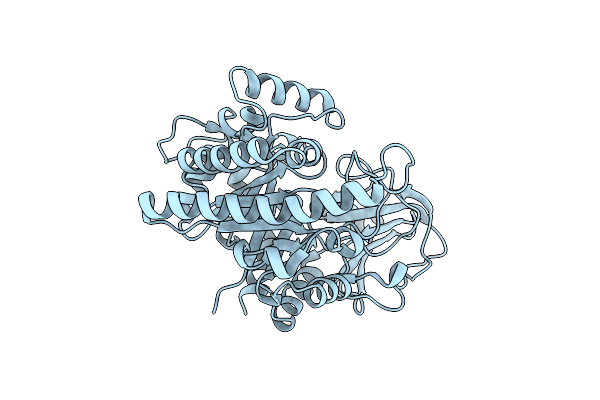 |
Organism: Vibrio parahaemolyticus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-09-25 Classification: HYDROLASE |
 |
Organism: Vibrio parahaemolyticus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-09-11 Classification: MEMBRANE PROTEIN |
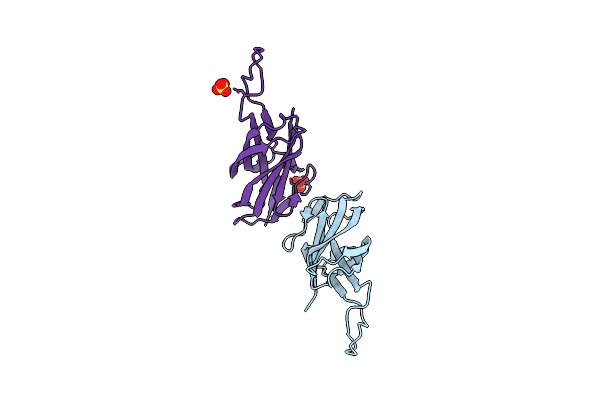 |
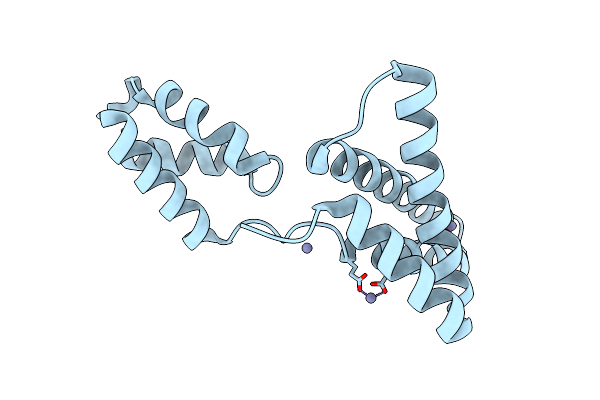
Intramolecular Ester Bond-Containing Repeat Domain From Gemella Massiliensis Adhesin
Organism: Gemella massiliensis
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2024-06-19 Classification: CELL ADHESION Ligands: SO4 |
 |
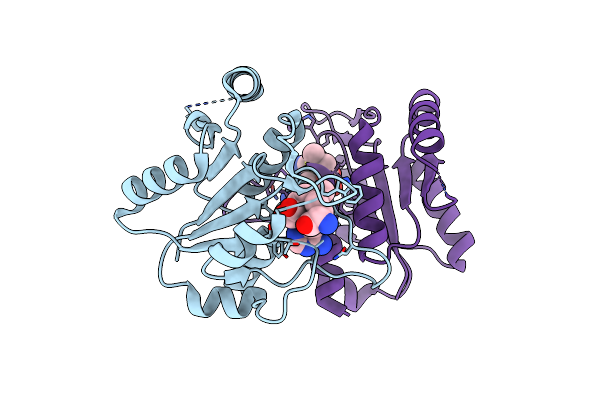
Organism: Vibrio parahaemolyticus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-05-22 Classification: UNKNOWN FUNCTION Ligands: ZN |
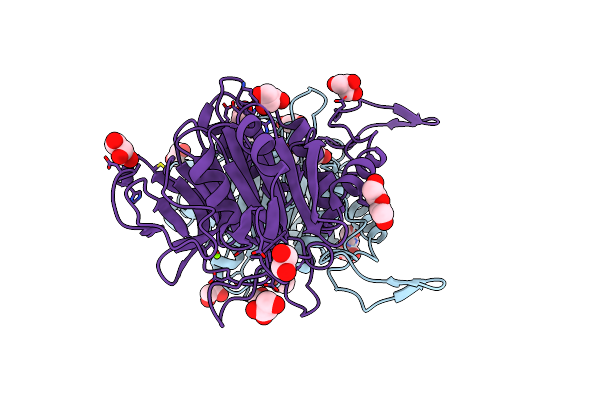 |
Organism: Arthrobacter citreus
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2024-02-07 Classification: HYDROLASE Ligands: MG, EDO, TRS, PEG, GOL |
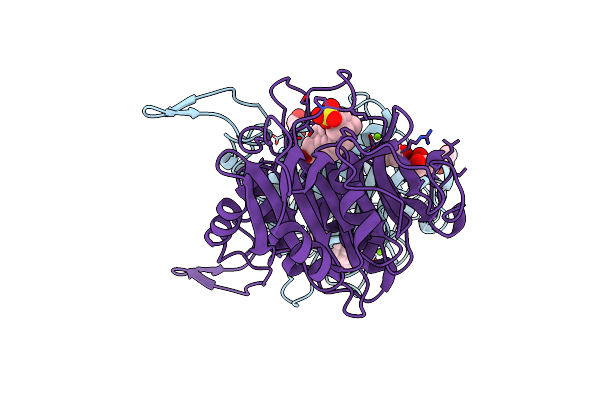 |
Bile Salt Hydrolase From Arthrobacter Citreus With Covalent Inhibitor Aaa-10 Bound
Organism: Arthrobacter citreus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-02-07 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: MG, CL, GOL, WSR |
 |
Crystal Structure Of Tm1570 Domain From Calditerrivibrio Nitroreducens In Complex With S-Adenosyl-L-Methionine
Organism: Calditerrivibrio nitroreducens dsm 19672
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-01-10 Classification: TRANSFERASE Ligands: SAM |
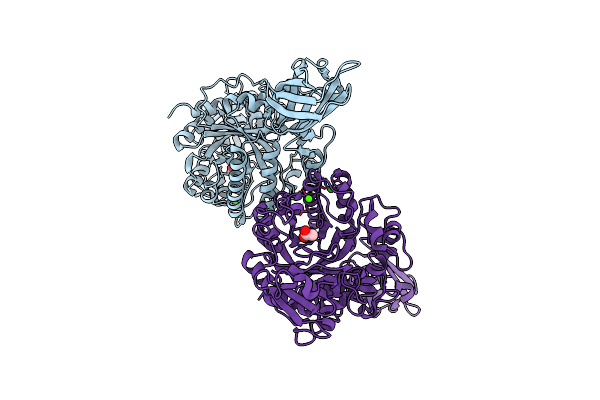 |
Organism: Vibrio parahaemolyticus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-12-20 Classification: HYDROLASE Ligands: ACT, CA |
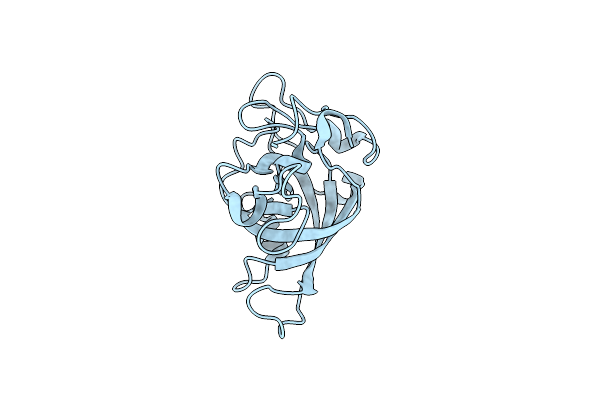 |
Organism: Gloeophyllum trabeum atcc 11539
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-11-22 Classification: HYDROLASE |
 |
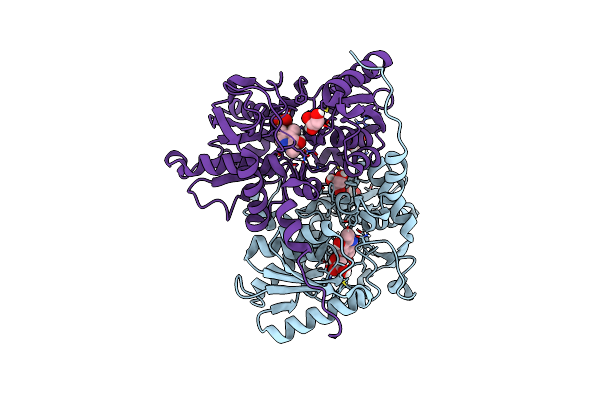
Ldh Mutant P101Q-(An Unexpected Single-Point Mutation Triggers The Unleashing Of Catalytic Potential Of A Nadh-Dependent Dehydrogenase)
Organism: Thermodesulfatator indicus dsm 15286
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-09-06 Classification: BIOSYNTHETIC PROTEIN |
 |
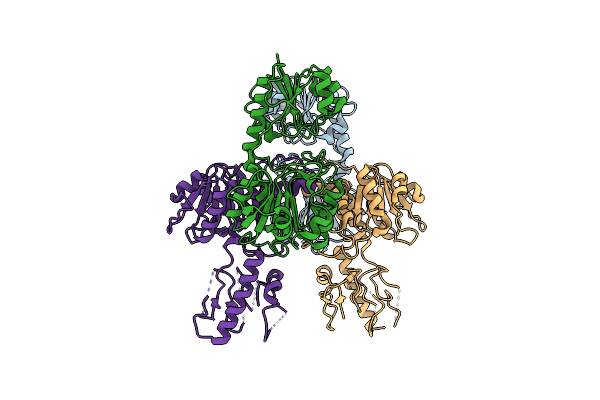
Vibrio Parahaemolyticus Vtra/Vtrc Complex Bound To The Bile Salt Chenodeoxycholate
Organism: Vibrio parahaemolyticus
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2023-06-14 Classification: SIGNALING PROTEIN Ligands: EDO, JN3, PG4, CA |
 |
Organism: Trypanosoma theileri
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2023-05-31 Classification: TRANSFERASE Ligands: PLP, GOL, CL |
 |
Crystal Structure Of Trmd Domain From Calditerrivibrio Nitroreducens In Complex With S-Adenosyl-L-Methionine
Organism: Calditerrivibrio nitroreducens dsm 19672
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2023-01-11 Classification: TRANSFERASE Ligands: EDO, GOL, CL, SAM |

