Search Count: 190
 |
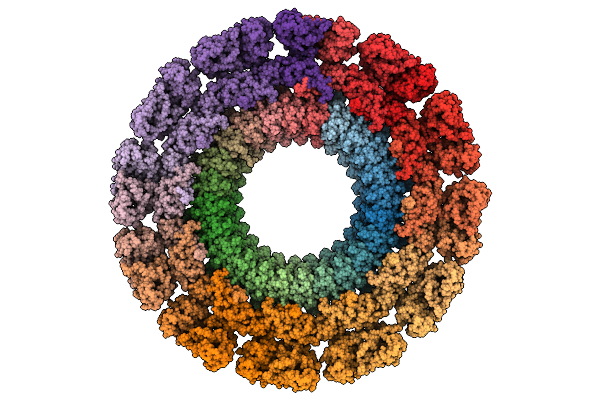
Cryo-Em Structure Of Gp77 Within The In Vitro Reconstituted Razr:Gp77 Complex
Organism: Escherichia phage secphi27
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN |
 |
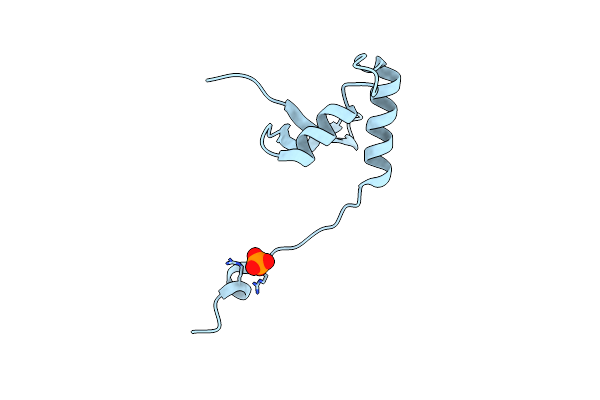
Cryo-Em Map Of The In Vitro Reconstituted Razr:Gp77 Complex With Alphafold-Predicted Models Fitted Into The Density.
Organism: Escherichia phage secphi27, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Pseudomonas phage luz7
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-10-30 Classification: VIRAL PROTEIN Ligands: PO4 |
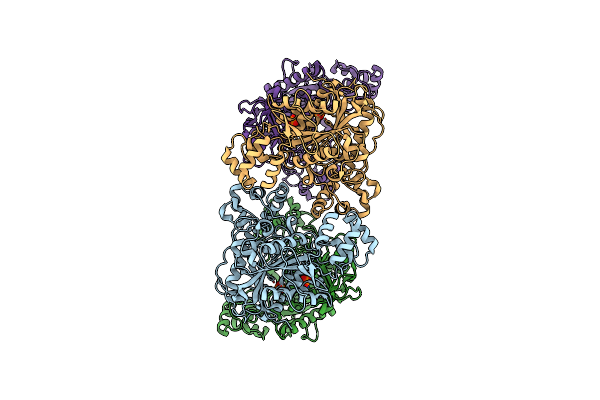 |
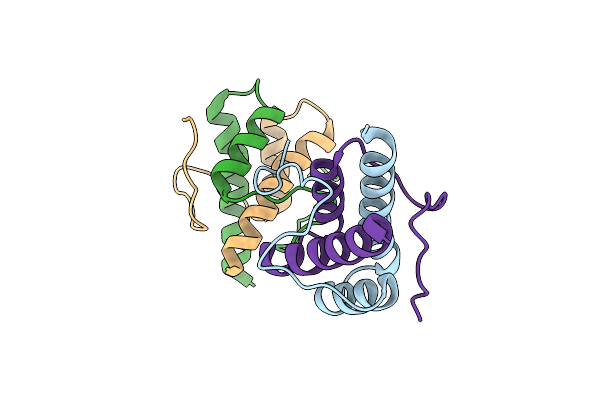
An Esterase From Anaerobic Clostridium Hathewayi Can Hydrolyze Aliphatic Aromatic Polyesters
Organism: Hungatella hathewayi
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-02-24 Classification: HYDROLASE Ligands: PO4 |
 |
The Structure Of Uncharacterized Protein Pp-Luz7_Gp033 From Pseudomonas Phage Luz7.
Organism: Pseudomonas phage luz7
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-11-10 Classification: structural genomics, unknown function Ligands: CL |
 |
Organism: Mytilus edulis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2006-02-22 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Mytilus edulis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-05-04 Classification: ISOMERASE |
 |
Organism: Mytilus edulis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-05-04 Classification: ISOMERASE Ligands: PEG |
 |
Organism: Mytilus edulis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-05-04 Classification: ISOMERASE Ligands: MG |
 |
Organism: Mytilus edulis
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2004-05-04 Classification: ISOMERASE Ligands: SO4 |
 |
Crystal Structure Of Phosphoenolpyruvate Mutase Complexed With Sulfopyruvate
Organism: Mytilus edulis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2002-08-28 Classification: ISOMERASE Ligands: MG, SPV |
 |
Organism: Mytilus edulis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1999-07-21 Classification: PHOSPHOTRANSFERASE Ligands: OXL, MG |
 |
 |
 |
 |
 |
 |
 |
 |

