Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
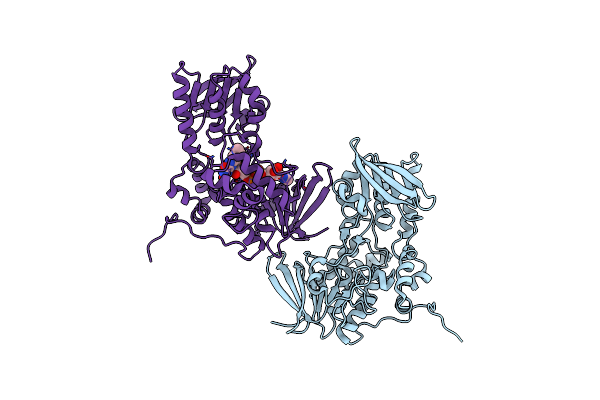 |
The Crystal Structure Of Dfoa Bound To Fad, The Desferrioxamine Biosynthetic Pathway Cadaverine Monooxygenase From The Fire Blight Disease Pathogen Erwinia Amylovora
Organism: Erwinia amylovora cfbp1430
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2018-06-27 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD |
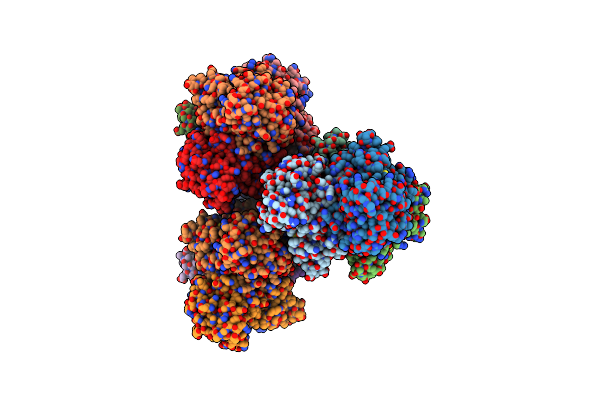 |
Crystal Structure Of Sorbitol-6-Phosphate 2-Dehydrogenase Srld From Erwinia Amylovora
Organism: Erwinia amylovora cfbp1430
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2018-06-13 Classification: OXIDOREDUCTASE Ligands: CL |
 |
The Crystal Structure Of Dfoj, The Desferrioxamine Biosynthetic Pathway Lysine Decarboxylase From The Fire Blight Disease Pathogen Erwinia Amylovora
Organism: Erwinia amylovora (strain cfbp1430)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-02-28 Classification: LYASE Ligands: PLP |
 |
The Crystal Structure Of Dfoc, The Desferrioxamine Biosynthetic Pathway Acetyltransferase/Non-Ribosomal Peptide Synthetase (Nrps)-Independent Siderophore (Nis) From The Fire Blight Disease Pathogen Erwinia Amylovora
Organism: Erwinia amylovora cfbp1430
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2018-02-28 Classification: BIOSYNTHETIC PROTEIN |
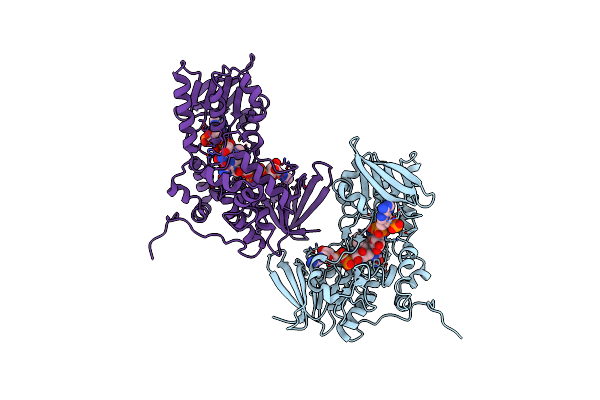 |
The Crystal Structure Of Dfoa Bound To Fad And Nadp; The Desferrioxamine Biosynthetic Pathway Cadaverine Monooxygenase From The Fire Blight Disease Pathogen Erwinia Amylovora
Organism: Erwinia amylovora cfbp1430
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-02-28 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, NAP |
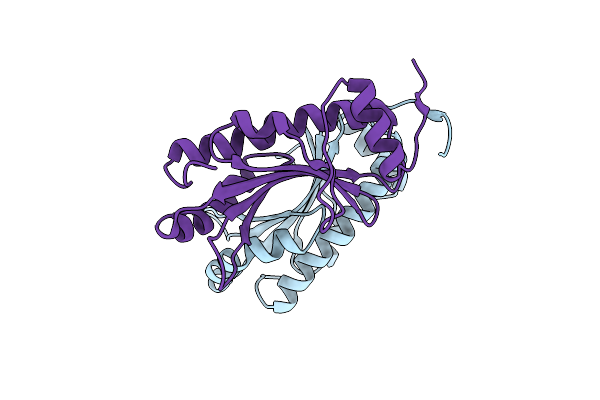 |
Semet Crystal Structure Of Erwinia Amylovora Amyr Amylovoran Repressor, A Member Of The Ybjn Protein Family
Organism: Erwinia amylovora
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2017-02-15 Classification: TRANSCRIPTION |
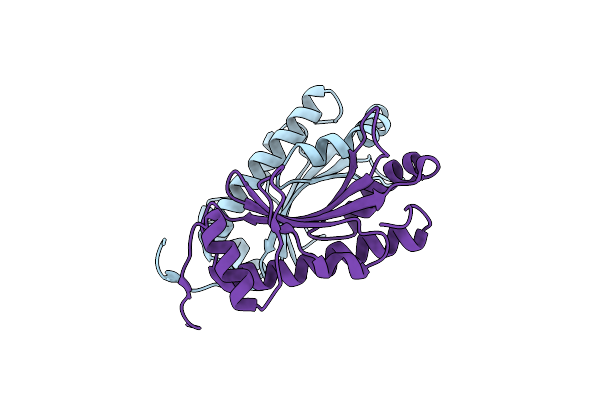 |
Erwinia Amylovora Amyr Amylovoran Repressor, A Member Of The Ybjn Protein Family
Organism: Erwinia amylovora
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-01-18 Classification: SIGNALING PROTEIN |
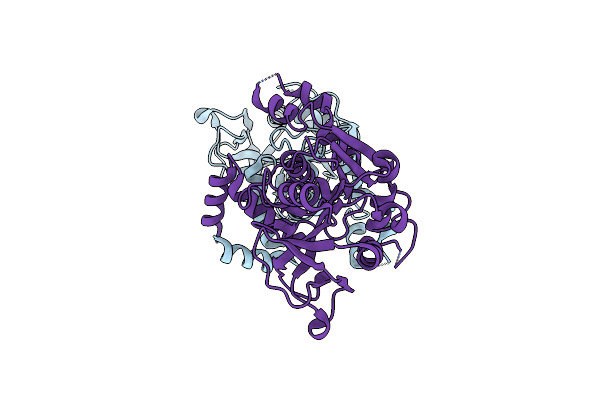 |
Crystal Structure Of Glucose-1-Phosphate Uridylyltransferase Galu From Erwinia Amylovora.
Organism: Erwinia amylovora
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2016-01-20 Classification: TRANSFERASE |
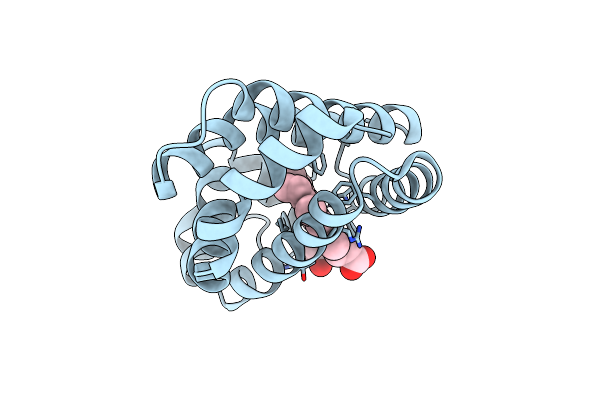 |
2.0 A Crystal Structure Of The Double Mutant H(E7)V, T(E10)R Of Myoglobin From Aplysia Limacina
Organism: Aplysia limacina
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2000-06-21 Classification: OXYGEN STORAGE/TRANSPORT Ligands: HEM |
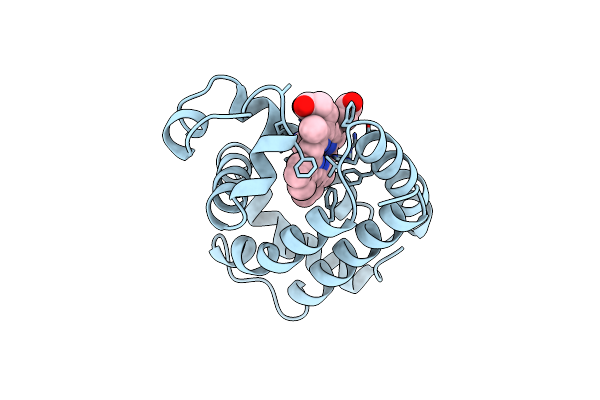 |
X-Ray Crystal Structure Of Ferric Aplysia Limacina Myoglobin In Different Liganded States
Organism: Aplysia limacina
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1993-10-31 Classification: OXYGEN STORAGE Ligands: CYN, HEM |
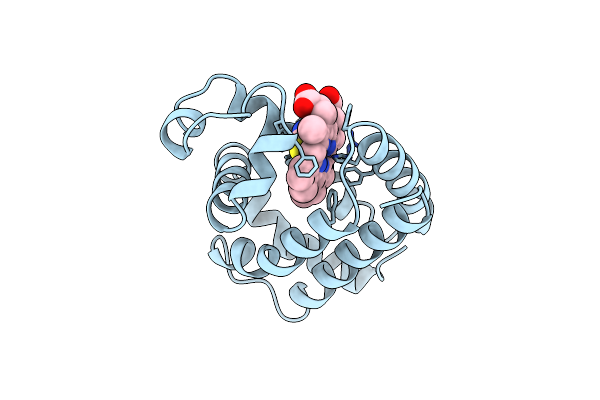 |
X-Ray Crystal Structure Of Ferric Aplysia Limacina Myoglobin In Different Liganded States
Organism: Aplysia limacina
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1993-10-31 Classification: OXYGEN STORAGE Ligands: SCN, HEM |
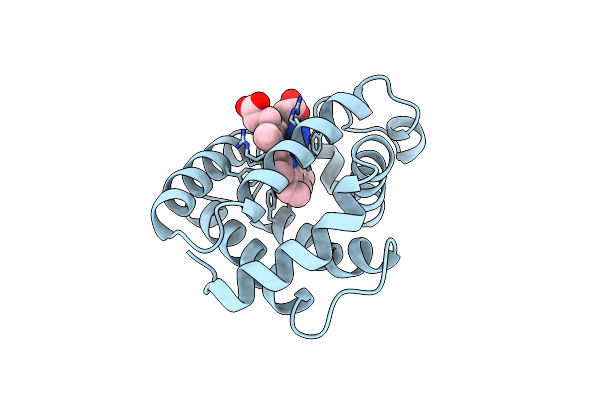 |
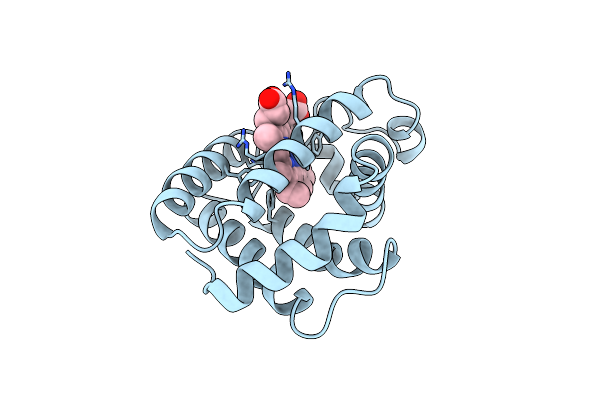
Binding Mode Of Azide To Ferric Aplysia Limacina Myoglobin. Crystallographic Analysis At 1.9 Angstroms Resolution
Organism: Aplysia limacina
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1992-07-15 Classification: OXYGEN STORAGE Ligands: AZI, HEM |
 |
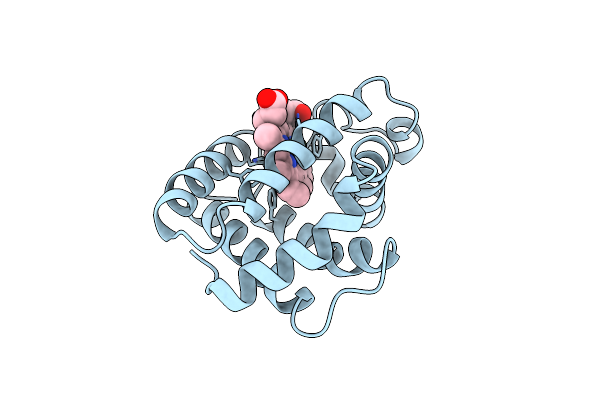
Aplysia Limacina Myoglobin. Crystallographic Analysis At 1.6 Angstroms Resolution
Organism: Aplysia limacina
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 1990-01-15 Classification: OXYGEN STORAGE Ligands: HEM |
 |
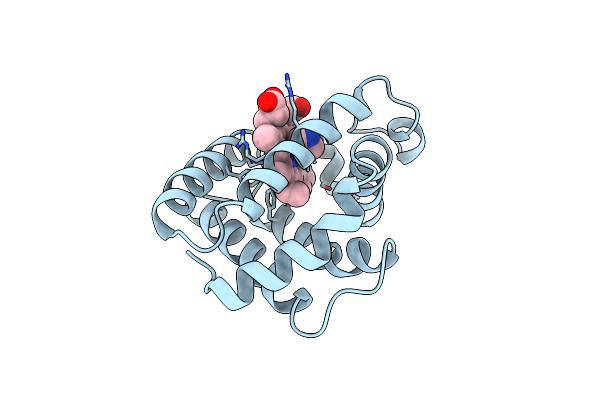
Aplysia Limacina Myoglobin. Crystallographic Analysis At 1.6 Angstroms Resolution
Organism: Aplysia limacina
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1990-01-15 Classification: OXYGEN STORAGE Ligands: F, HEM |
 |
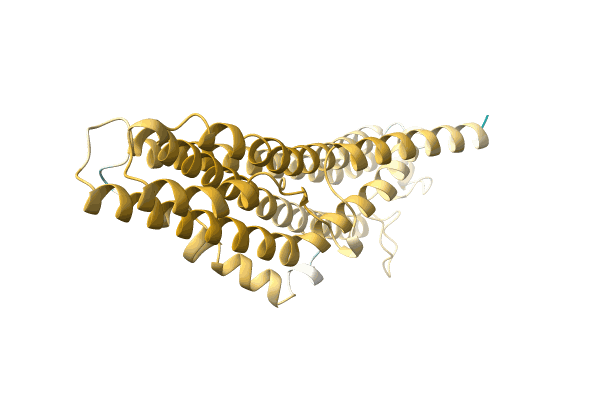
Aplysia Limacina Myoglobin. Crystallographic Analysis At 1.6 Angstroms Resolution
Organism: Aplysia limacina
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1990-01-15 Classification: OXYGEN STORAGE Ligands: HEM, IMD |
 |
 |
 |
 |
 |