Search Count: 2,742
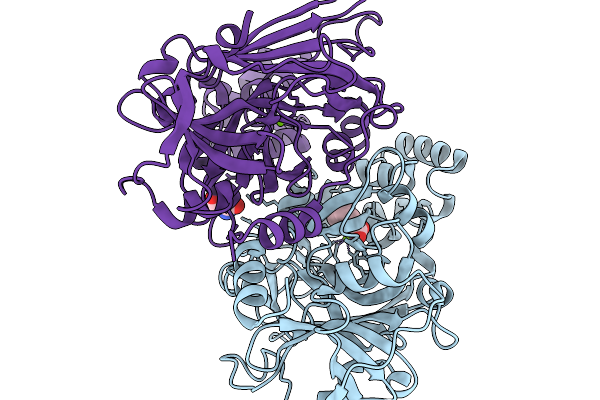 |
Structure-Guided Interface Engineering For Modifying Substrate Binding And Catalytic Activity Of 2-Keto-3-Deoxy-D-Xylonate Dehydratase
Organism: Caulobacter vibrioides na1000
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: CARBOHYDRATE Ligands: MG, TRS, COI |
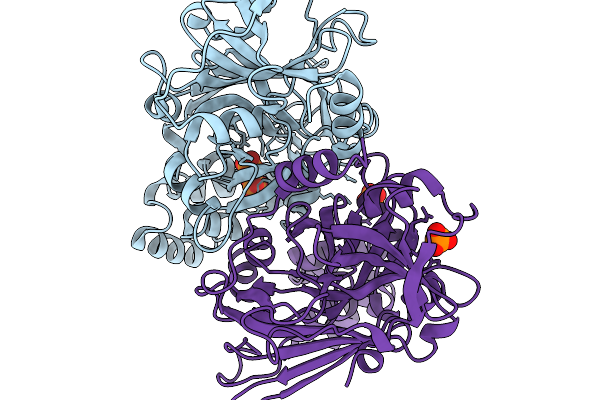 |
Structure-Guided Interface Engineering For Modifying Substrate Binding And Catalytic Activity Of 2-Keto-3-Deoxy-D-Xylonate Dehydratase
Organism: Caulobacter vibrioides na1000
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: CARBOHYDRATE Ligands: 2HP, NH4 |
 |
Organism: Aplysia californica
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: HYDROLASE Ligands: NAG, ZN |
 |
Chimeric Adenosine Deaminase Growth Factor (Adgf) In Complex With Pentostatin
Organism: Aplysia californica, Helix pomatia
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: HYDROLASE Ligands: NAG, EDO, ZN, DCF |
 |
Organism: Aplysia californica
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: MEMBRANE PROTEIN Ligands: K, MG, CA |
 |
Organism: Aplysia californica
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: MEMBRANE PROTEIN Ligands: K |
 |
Organism: Aplysia californica
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: MEMBRANE PROTEIN Ligands: K, MG, CA |
 |
Organism: Aplysia californica
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: MEMBRANE PROTEIN Ligands: K |
 |
Organism: Aplysia californica
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: MEMBRANE PROTEIN Ligands: K, MG, CA |
 |
Organism: Aplysia californica
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: MEMBRANE PROTEIN Ligands: CA, K |
 |
Chimeric Adenosine Deaminase Growth Factor (Adgf) In Complex With Pentostatin Monophosphate
Organism: Aplysia californica
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: HYDROLASE Ligands: ZN, A1CDW, NAG, EDO, GOL |
 |
Organism: Aplysia californica
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: MEMBRANE PROTEIN |
 |
Organism: Coronavirus hku15
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2025-05-21 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens, Vicugna pacos, Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:3.28 Å Release Date: 2025-05-21 Classification: OXIDOREDUCTASE/IMMUNE SYSTEM Ligands: ZN |
 |
Organism: Caulobacter vibrioides cb15
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: DNA BINDING PROTEIN Ligands: GOL, SO4 |
 |
Organism: Caulobacter vibrioides
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MOTOR PROTEIN |
 |
Organism: Caulobacter vibrioides
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MOTOR PROTEIN |
 |
Compact Structure Of Cpaf With Two Atps And Two Adps (Under-Saturated Atp/Adp Dataset)
Organism: Caulobacter vibrioides
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MOTOR PROTEIN Ligands: MG, ATP, ADP |
 |
Expanded Structure Of Cpaf With Two Atps And Four Adps (Under-Saturated Atp/Adp Dataset)
Organism: Caulobacter vibrioides
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MOTOR PROTEIN Ligands: MG, ATP, ADP |
 |
Expanded Structure Of Cpaf With Two Atps And Four Adps (Saturated Atp Dataset)
Organism: Caulobacter vibrioides
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MOTOR PROTEIN Ligands: MG, ATP, ADP |

