Search Count: 45
 |
Organism: Streptomyces sp. cfmr 7
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: ACY, MG, EDO |
 |
Crystal Structure Of Hrmj From Streptomyces Sp. Cfmr 7 (Hrmj-Ssc) Complexed With Manganese (Ii), 2-Oxoglutarate And 6-Nitronorleucine
Organism: Streptomyces sp. cfmr 7
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: MN, AKG, 6HN, IOD, EDO, GOL |
 |
Crystal Structure Of Hrmj From Streptomyces Sp. Cfmr 7 (Hrmj-Ssc) Complexed With Vanadyl(Iv)-Oxo, Succinate And 6-Nitronorleucine
Organism: Streptomyces sp. cfmr 7
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: 6HN, VVO, SIN, GOL, BR |
 |
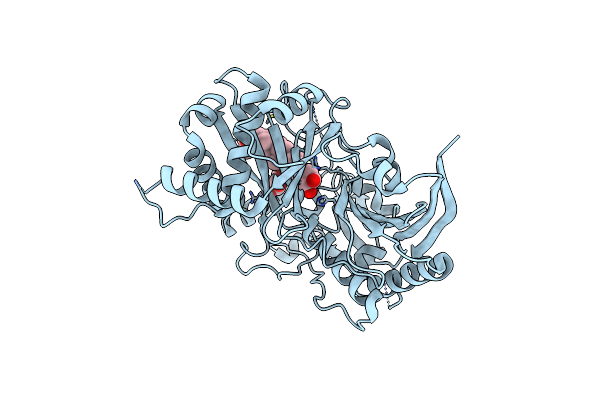
Organism: Mammarenavirus lujoense
Method: ELECTRON MICROSCOPY Release Date: 2024-06-12 Classification: VIRAL PROTEIN Ligands: NAG, NA |
 |
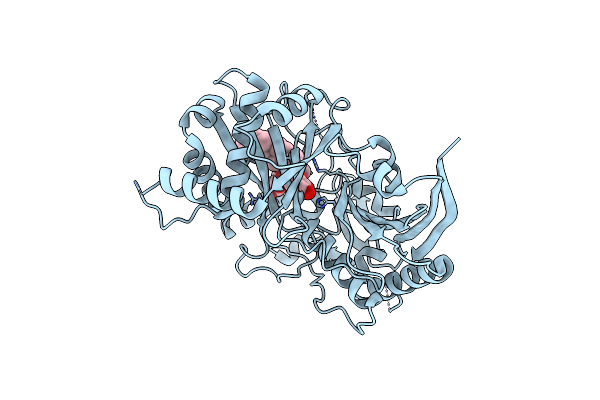
Organism: Astragalus membranaceus
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2023-09-13 Classification: BIOSYNTHETIC PROTEIN Ligands: KZO |
 |
Organism: Astragalus membranaceus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2023-09-13 Classification: BIOSYNTHETIC PROTEIN Ligands: KZO |
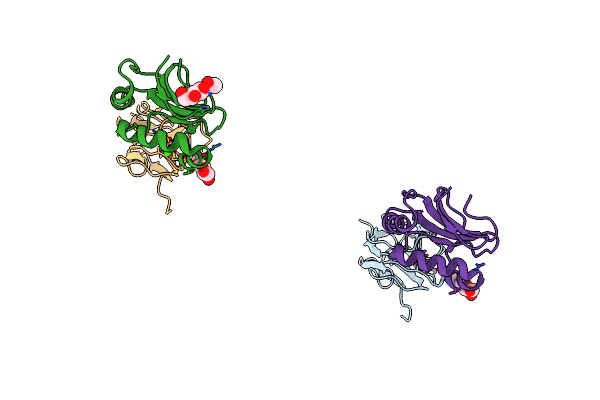 |
Crystal Structure Of Gp1 Domain Of Lujo Virus In Complex With The First Cub Domain Of Neuropilin-2
Organism: Lujo mammarenavirus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2018-07-25 Classification: VIRAL PROTEIN Ligands: NAG, CA |
 |
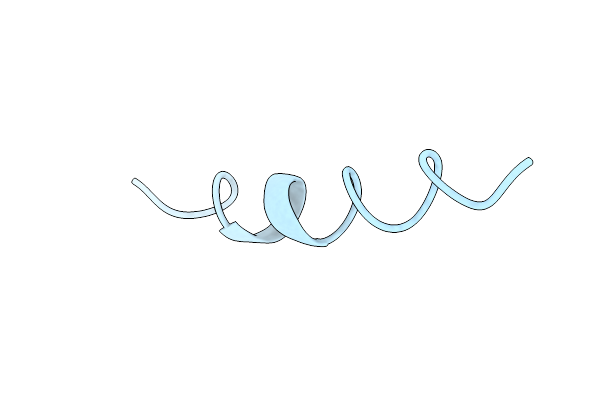
Structure Of The Calcium Channel Blocker Omega Conotoxin Gvia, Nmr, 20 Structures
|
 |
 |
Nmr Structure Of Calcium Bound Conformer Of Conantokin G, Minimized Average Structure
|
 |
 |
Organism: Conus geographus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 1996-12-07 Classification: CONOTOXIN |
 |
 |
Solution Structure Of Omega-Conotoxin Gvia Using 2-D Nmr Spectroscopy And Relaxation Matrix Analysis
Organism: Conus geographus
Method: SOLUTION NMR Release Date: 1994-01-31 Classification: PRESYNAPTIC NEUROTOXIN |
 |
Structure-Activity Relationships Of Mu-Conotoxin Giiia: Structure Determination Of Active And Inactive Sodium Channel Blocker Peptides By Nmr And Simulated Annealing Calculations
|
 |
Structure-Activity Relationships Of Mu-Conotoxin Giiia: Structure Determination Of Active And Inactive Sodium Channel Blocker Peptides By Nmr And Simulated Annealing Calculations
|
 |
 |
 |
 |

