Search Count: 12
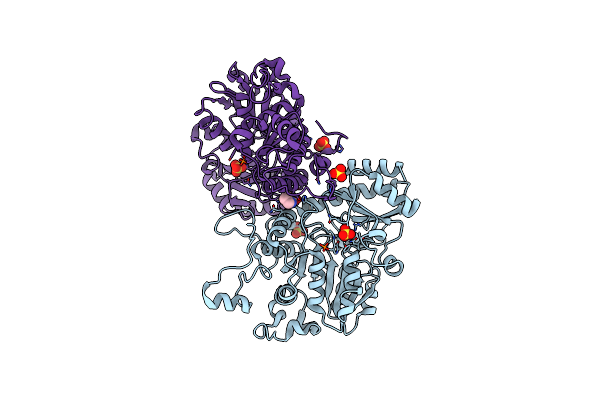 |
Crystal Structure Of O-Acetylhomoserine Sulfhydrylase From Lactobacillus Plantarum In The Closed Form
Organism: Lactiplantibacillus plantarum jdm1
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2024-02-14 Classification: LYASE Ligands: SO4, PRO |
 |
Crystal Structure Of O-Acetylhomoserine Sulfhydrylase From Lactobacillus Plantarum In The Open Form
Organism: Lactiplantibacillus plantarum jdm1
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-02-14 Classification: LYASE Ligands: PRO |
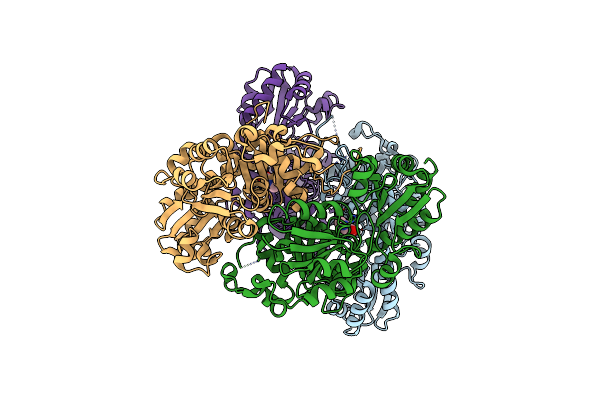
 |
Organism: Lactobacillus plantarum jdm1
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2016-05-04 Classification: TRANSFERASE Ligands: ADP |
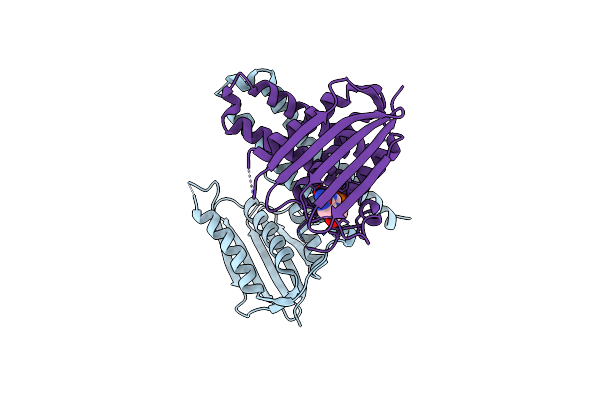
 |
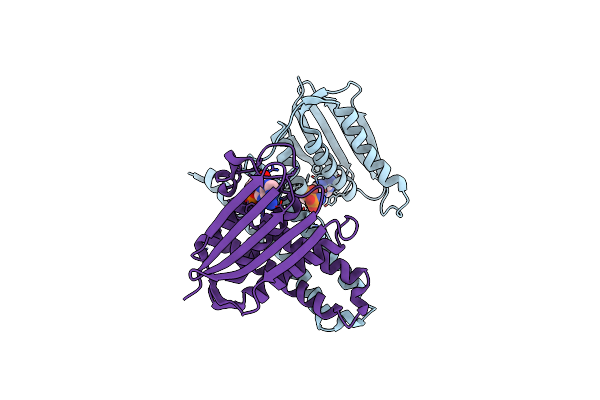
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2015-09-02 Classification: TRANSFERASE |
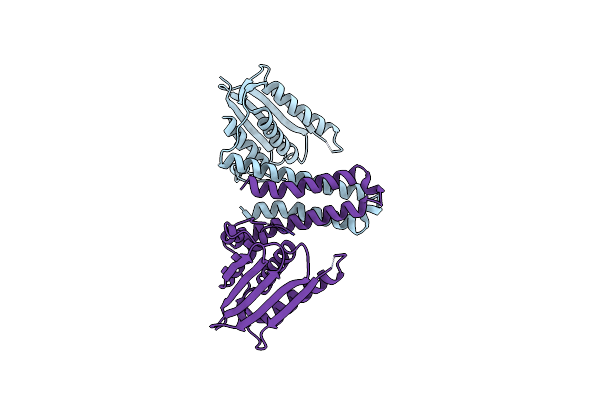
 |
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-09-02 Classification: TRANSFERASE Ligands: AN2 |
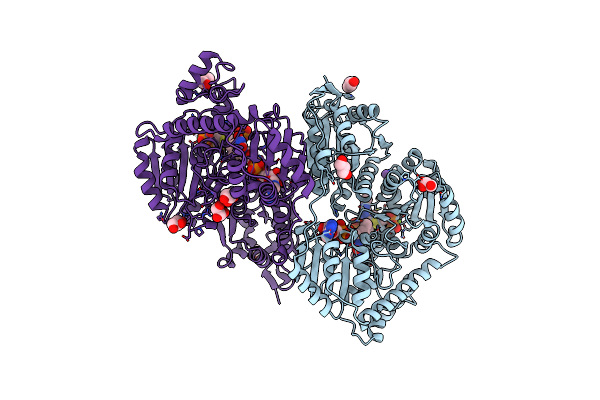 |
High-Resolution Crystal Structure Of Pyruvate Oxidase From L. Plantarum In Complex With Phosphate
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2013-06-12 Classification: OXIDOREDUCTASE Ligands: FAD, MG, PO4, TPP, GOL, K |
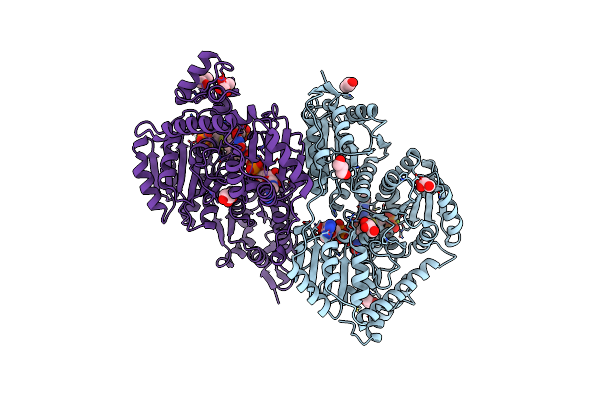 |
High-Resolution Structure Of Pyruvate Oxidase In Complex With Reaction Intermediate 2-Hydroxyethyl-Thiamin Diphosphate Carbanion-Enamine, Crystal B
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2012-06-20 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, PYR, PO4, TDM, MG |
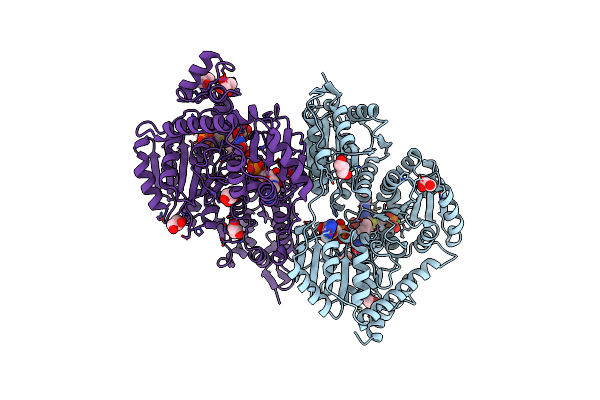 |
High-Resolution Structure Of Pyruvate Oxidase In Complex With Reaction Intermediate 2-Hydroxyethyl-Thiamin Diphosphate Carbanion-Enamine, Crystal A
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2012-06-20 Classification: OXIDOREDUCTASE Ligands: PYR, PO4, GOL, MG, TDM, FAD |
 |
NA
Organism: Lactobacillus plantarum (strain JDM1)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Lactobacillus plantarum (strain JDM1)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Lactobacillus plantarum (strain JDM1)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Lactobacillus plantarum (strain JDM1)
Method: Alphafold Release Date: Classification: NA Ligands: NA |

