Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Syntrophothermus lipocalidus (strain dsm 12680 / tgb-c1)
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2024-07-17 Classification: OXIDOREDUCTASE Ligands: NAI, SO4, MG, EDO, TRS |
 |
Organism: Larkinella arboricola
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2024-01-03 Classification: SIGNALING PROTEIN Ligands: C2E |
 |
Organism: Syntrophothermus lipocalidus dsm 12680
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-11-14 Classification: RNA Ligands: PRP, MN, NA, K |
 |
Organism: Syntrophothermus lipocalidus dsm 12680
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2018-11-14 Classification: RNA Ligands: PRP, IRI, MG, ACT, K |
 |
Organism: Syntrophothermus lipocalidus dsm 12680
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2018-11-14 Classification: RNA Ligands: PRP, TL, MG |
 |
Organism: Syntrophothermus lipocalidus dsm 12680
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-11-14 Classification: RNA Ligands: MG, NA, PRP |
 |
Organism: Syntrophothermus lipocalidus dsm 12680
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-11-14 Classification: RNA Ligands: MG |
 |
Structural Basis For The Function Of Tim50 In The Mitochondrial Presequence Translocase
Organism: Saccharomyces cerevisiae ec1118
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2011-03-02 Classification: CHAPERONE Ligands: CA, ACT, 1PE |
 |
Organism: Themiste dyscritum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1992-01-15 Classification: OXYGEN TRANSPORT Ligands: ACE, FEO |
 |
Organism: Themiste dyscritum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1992-01-15 Classification: OXYGEN TRANSPORT Ligands: ACE, FEO, OXY |
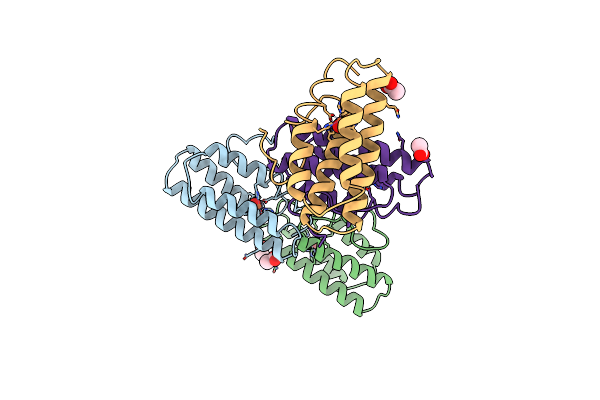 |
The Structures Of Met And Azidomet Hemerythrin At 1.66 Angstroms Resolution
Organism: Themiste dyscritum
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 1992-01-15 Classification: OXYGEN TRANSPORT Ligands: ACT, FEO |
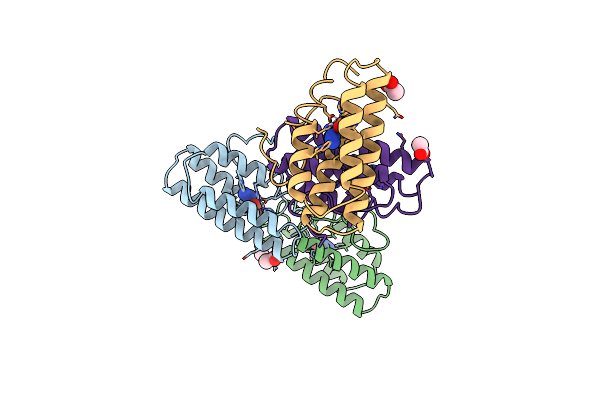 |
The Structures Of Met And Azidomet Hemerythrin At 1.66 Angstroms Resolution
Organism: Themiste dyscritum
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 1992-01-15 Classification: OXYGEN TRANSPORT Ligands: ACT, FEA |
 |
 |
 |
 |
 |
 |
 |
 |