Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Saccharomyces cerevisiae bmn1-35, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RNA BINDING PROTEIN/RNA Ligands: AN6 |
 |
Organism: Human alphaherpesvirus 1 strain r-15
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: VIRAL PROTEIN Ligands: ZN, A1BXB |
 |
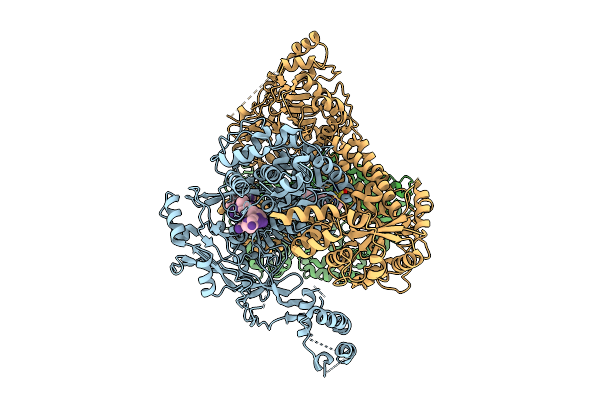
Organism: Human alphaherpesvirus 1 strain r-15
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: VIRAL PROTEIN Ligands: ZN, A1CHB |
 |
Organism: Human alphaherpesvirus 1 strain r-15
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: VIRAL PROTEIN Ligands: ZN, A1BXB |
 |
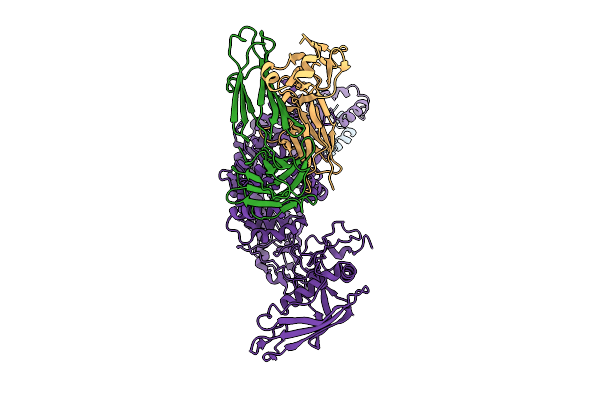
Organism: Human alphaherpesvirus 1 strain r-15
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: VIRAL PROTEIN Ligands: ZN, A1BXD |
 |
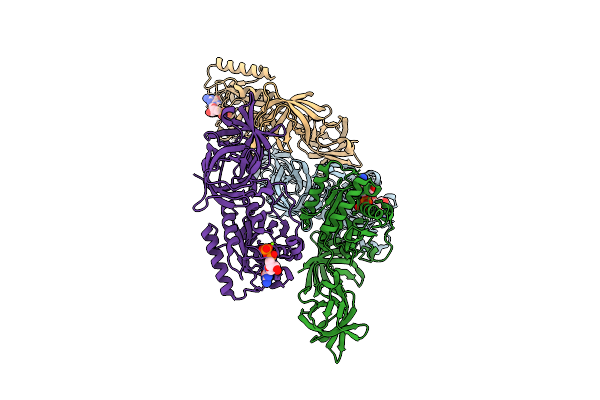
P116 From Mycoplasma Pneumoniae In Complex With Mild Growth Suppressor Monoclonal Antibody
Organism: Mus musculus, Mycoplasmoides pneumoniae m129
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: PROTEIN TRANSPORT |
 |
Organism: Mycoplasma pneumoniae (strain atcc 29342 / m129 / subtype 1)
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: BIOSYNTHETIC PROTEIN Ligands: GDP, MG |
 |
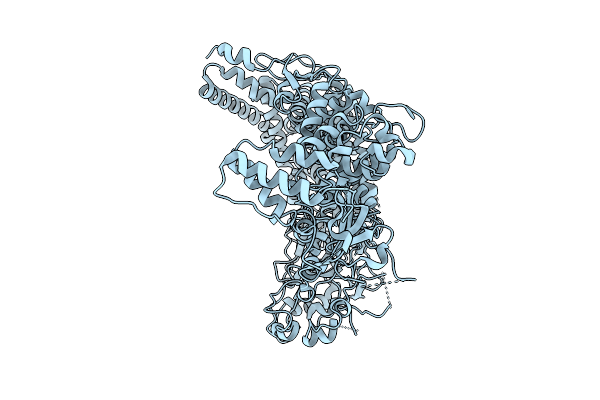
Cryo-Em Structure Of Essential Mycoplasma Pneumoniae Lipoprotein Mpn444 Homotrimer
Organism: Mycoplasmoides pneumoniae m129
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: UNKNOWN FUNCTION |
 |
Organism: Mycoplasmoides pneumoniae m129
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: UNKNOWN FUNCTION |
 |
Organism: Methanobrevibacter ruminantium m1
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: HYDROLASE |
 |
Organism: Methanobrevibacter ruminantium m1
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2025-04-23 Classification: HYDROLASE |
 |
Single-Particle Cryo-Em Of Mycoplasma Pneumoniae Adhesin P1 Complexed With The Anti-Adhesive Fab Fragment.
Organism: Mycoplasmoides pneumoniae m129, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: CELL ADHESION |
 |
Self Assembly Domain Of The Surface Layer Protein Of Viridibacillus Arvi (Aa 765-844)
Organism: Viridibacillus arvi
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-08-28 Classification: STRUCTURAL PROTEIN Ligands: BR |
 |
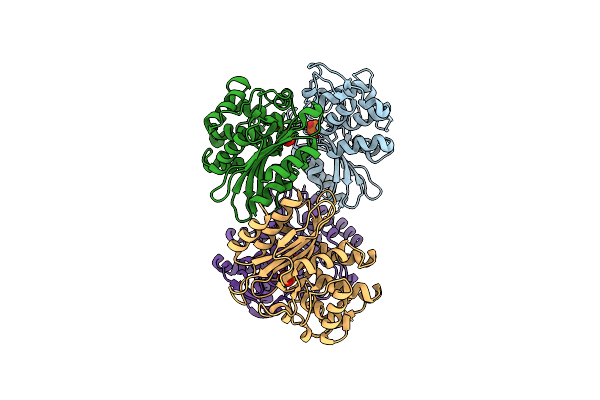
P116 Dimer In The Full State (Pdb Structure Of The Full-Length Ectodomain Truncated To Amino Acids 246-818)
Organism: Mycoplasmoides pneumoniae m129
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: LIPID BINDING PROTEIN |
 |
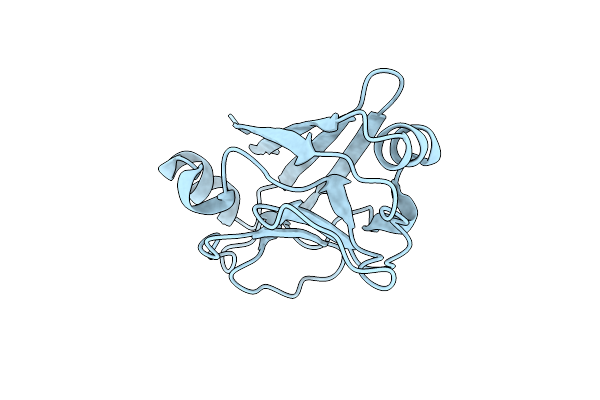
Xfel Structure Of Mycobacterium Tuberculosis Beta Lactamase Microcrystals Mixed With Sulbactam For 3 Ms
Organism: Mycobacterium tuberculosis str. beijing/w bt1
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2023-09-20 Classification: HYDROLASE/Inhibitor Ligands: PO4 |
 |
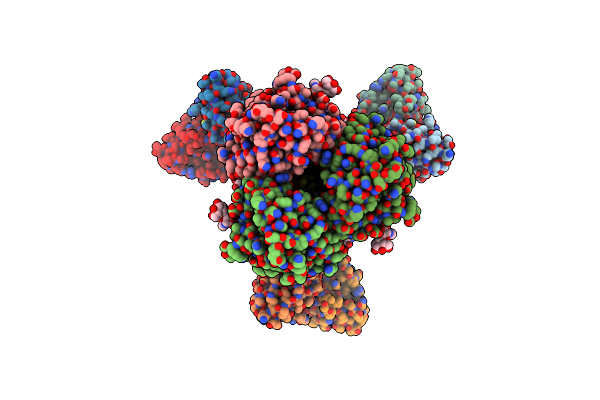
Crystal Structure Of Duf371 Domain-Containing Protein From Methanobrevibacter Ruminantium
Organism: Methanobrevibacter ruminantium
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2023-07-12 Classification: STRUCTURAL GENOMICS |
 |
Cryoem Structure Of Influenza A Virus A/Melbourne/1/1946 (H1N1) Hemagglutinin Bound To Cr6261 Fab
Organism: Influenza a virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Serratia plymuthica 4rx13
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-03-08 Classification: BIOSYNTHETIC PROTEIN Ligands: FE, GOL |
 |
Single Particle Cryo-Em Of The Lipid Binding Protein P116 (Mpn213) From Mycoplasma Pneumoniae At 3.3 Angstrom Resolution.
Organism: Mycoplasma pneumoniae m129
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: LIPID BINDING PROTEIN |
 |
Single Particle Cryo-Em Of The Empty Lipid Binding Protein P116 (Mpn213) From Mycoplasma Pneumoniae At 4 Angstrom Resolution
Organism: Mycoplasma pneumoniae m129
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: LIPID BINDING PROTEIN |