Search Count: 1,681
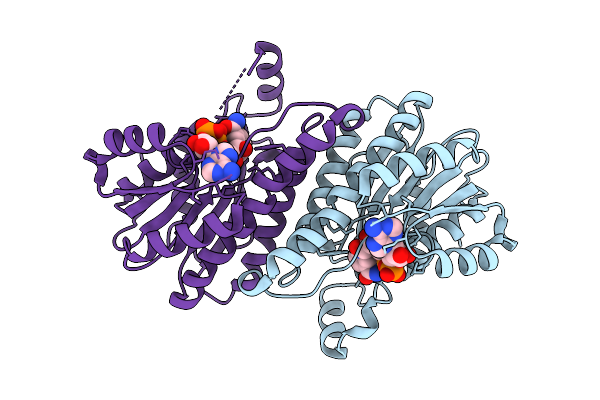 |
Crystal Structure Of L-2-Keto-3-Deoxypentonate 4-Dehydrogenase Bound To Nad(H)
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: OXIDOREDUCTASE Ligands: NAD |
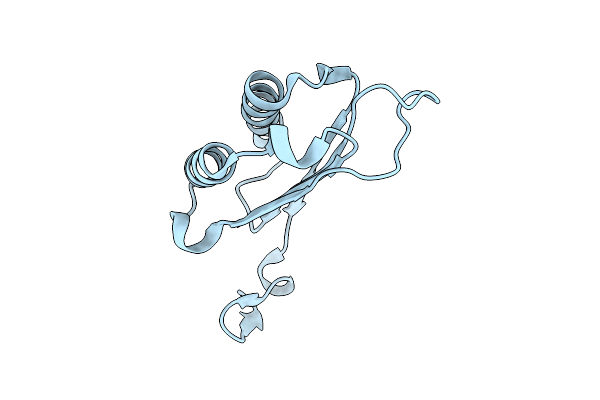 |
Organism: Heligmosomoides polygyrus
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: CYTOKINE |
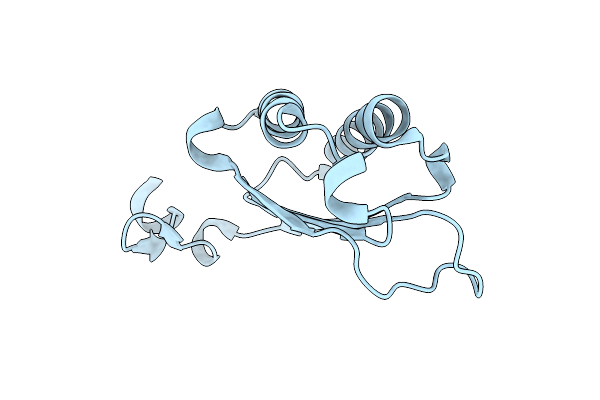 |
Organism: Heligmosomoides polygyrus
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: CYTOKINE |
 |
Organism: Actinia fragacea
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: MEMBRANE PROTEIN Ligands: FO4, CLR |
 |
Crysral Structure Of 2-Keto-3-Deoxypentonate 4-Dehydrogenase From Herbaspirillum Huttiense (Apo Form)
Organism: Herbaspirillum huttiense
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE |
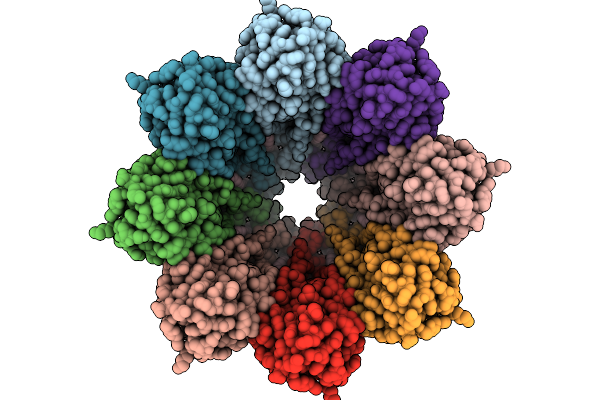 |
Structure Of The Octameric Pore Of Fragaceotxin C (Frac Or Delta-Actitoxin-Afr1A) In Large Unilamellar Vesicles.
Organism: Actinia fragacea
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: MEMBRANE PROTEIN Ligands: FO4, CLR |
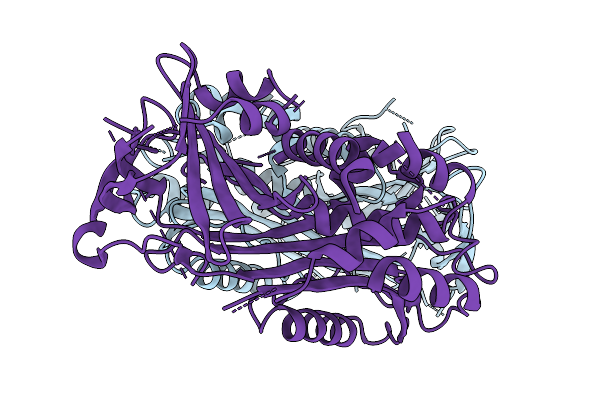 |
Structures And Mechanisms Of Serine Protease Inhibitors Of Trichinella Spiralis And Trichinella Pseudospiralis
Organism: Trichinella spiralis
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: STRUCTURAL PROTEIN |
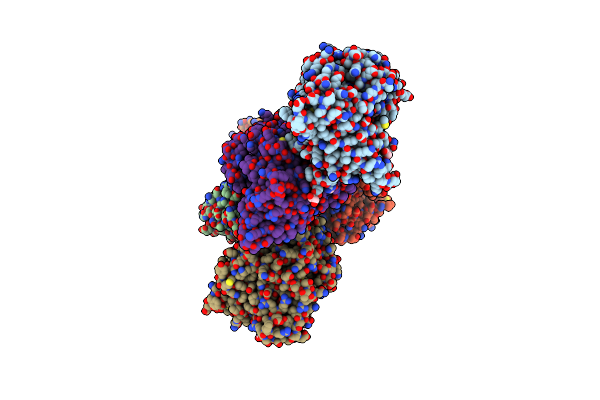 |
Organism: Lysinibacillus composti
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2025-04-09 Classification: OXIDOREDUCTASE Ligands: EDO, NAD |
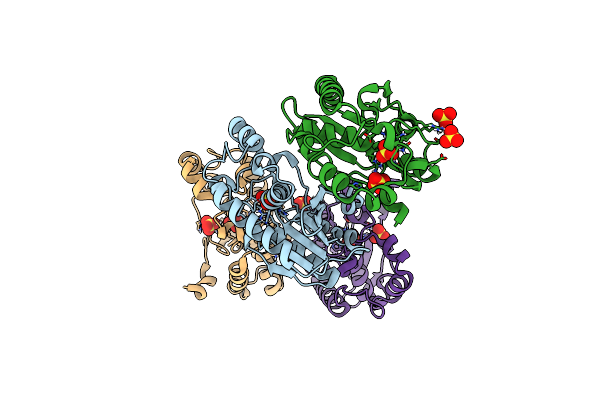 |
Hfp (Histidine Family Phosphatase) That Catalyzes Essential Dephosphorylation For Riboflavin Biosynthesis
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2025-04-09 Classification: HYDROLASE Ligands: SO4 |
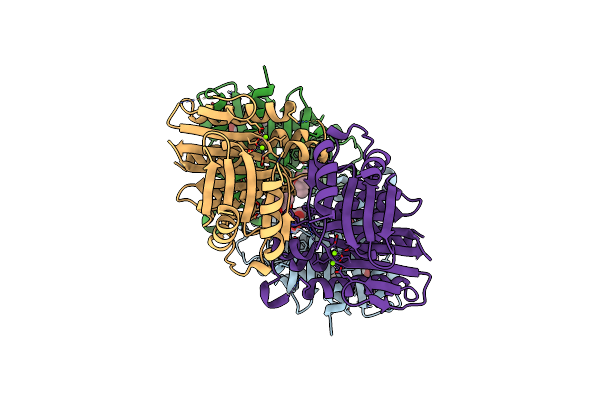 |
Myo-Inositol-1(Or 4)-Monophosphatase That Can Perform Essential Dephosphorylation Step To Facilitate Riboflavin Biosynthesis
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-04-09 Classification: HYDROLASE Ligands: MPD, MG, PO4 |
 |
Organism: Heligmosomoides polygyrus
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2025-03-05 Classification: STRUCTURAL PROTEIN |
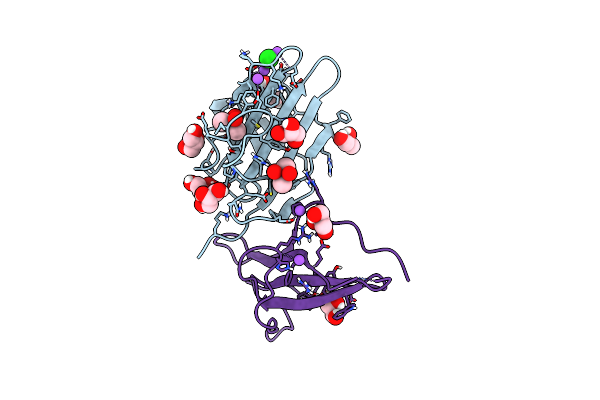 |
Heligmosomoides Polygyrus Tgf-Beta Mimic 6 Domain 3 (Tgm6-D3) Bound To Human Tgf-Beta Type Ii Receptor Extracellular Domain
Organism: Homo sapiens, Heligmosomoides polygyrus
Method: X-RAY DIFFRACTION Release Date: 2025-01-22 Classification: IMMUNOSUPPRESSANT Ligands: GOL, CL, NA, CAC |
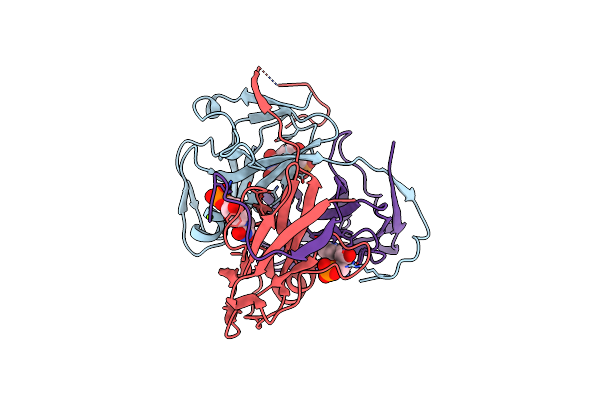 |
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: DU, MG |
 |
Organism: Dragon grouper nervous necrosis virus
Method: SOLUTION NMR Release Date: 2024-10-30 Classification: VIRAL PROTEIN |
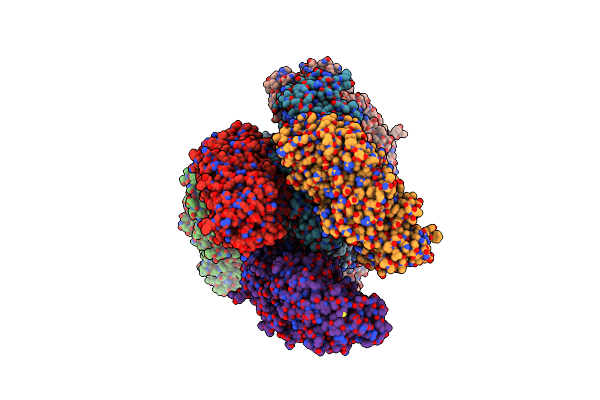 |
Crystal Structure Of 1,4-Alpha-Glucan Branching Protein From Rhodothermus Profundi
Organism: Rhodothermus profundi
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2024-10-02 Classification: BIOSYNTHETIC PROTEIN |
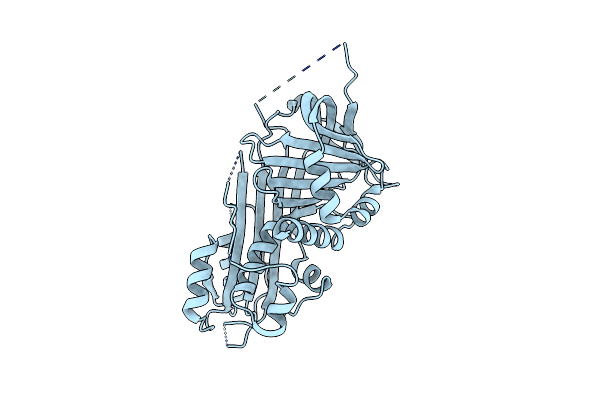 |
Structures And Mechanisms Of Serine Protease Inhibitors Of Trichinella Spiralis And Trichinella Pseudospiralis
Organism: Trichinella pseudospiralis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-09-25 Classification: STRUCTURAL PROTEIN |
 |
Organism: Micromonospora maris ab-18-032
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-09-18 Classification: LIGASE Ligands: EPE |
 |
Cryo-Em Structure Of Dragon Grouper Nervous Necrosis Virus-Like Particle At Ph8.0 (3.23A)
Organism: Dragon grouper nervous necrosis virus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: VIRUS Ligands: CA |
 |
Cryo-Em Structure Of Dragon Grouper Nervous Necrosis Virus-Like Particle At Ph6.5 (2.82A)
Organism: Dragon grouper nervous necrosis virus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: VIRUS Ligands: CA |
 |
Cryo-Em Structure Of Dragon Grouper Nervous Necrosis Virus-Like Particle At Ph5.0 (3.52A)
Organism: Dragon grouper nervous necrosis virus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: VIRUS Ligands: CA |

