Search Count: 4,080
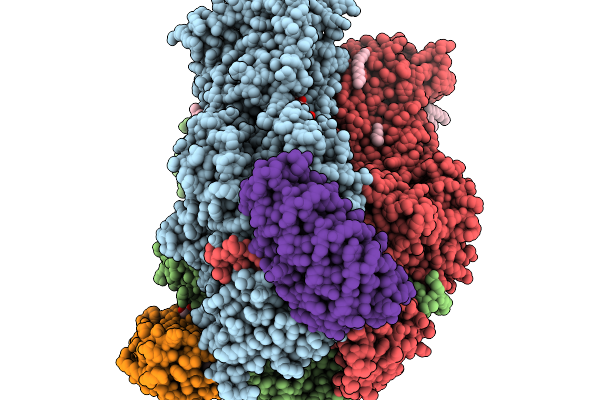 |
Structure Of The Double Cys-Substituted Cross-Linked Acrb Variant S562C_T837C
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TRANSPORT PROTEIN Ligands: LMT, D10, GOL, D12, HEX, C14, OCT, DD9, SO4 |
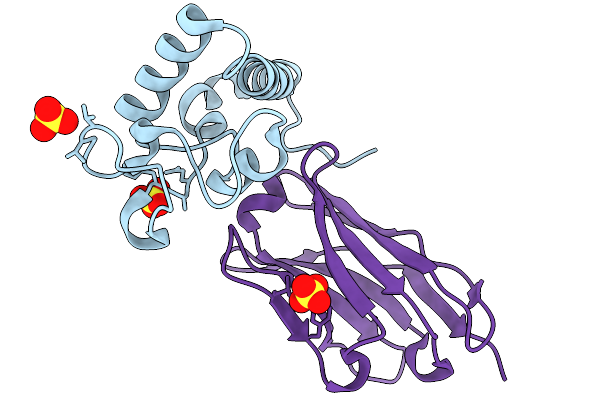 |
Crystal Structure Of Tetraspanin Cd63Mutant Large Extracellular Loop (Lel) In Complex With Sybody Sb3
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: CELL ADHESION Ligands: SO4 |
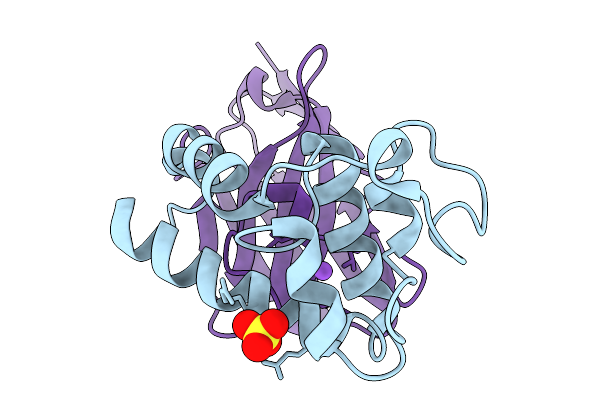 |
Crystal Structure Of Tetraspanin Cd63Mutant Large Extracellular Loop (Lel) In Complex With Sybody La4
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: CELL ADHESION Ligands: SO4, NA |
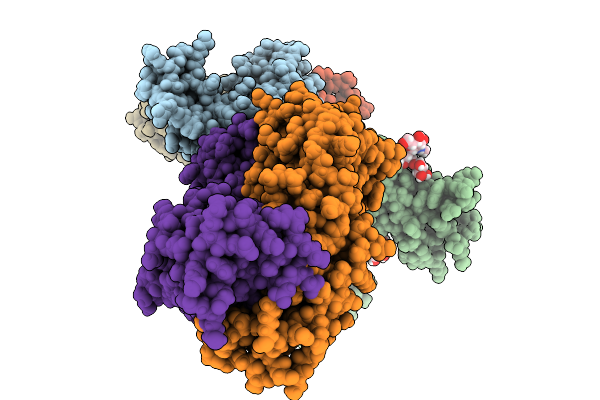 |
Cryo-Em Structure Of The Heterotrimeric Interleukin-2 Receptor In Complex With Interleukin-2 And Anti-Cd25 Fab S417
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: CIT, EDO |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: CU, CL, IMD, EDO, GOL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: NI, IMD, GOL, NA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN, IMD, EDO, GOL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: CO, IMD, GOL, NA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: TB, EDO, GOL, NA |
 |
Cryo-Em Structure Of Pyrene-Modified Tip60 Double Mutant (G12C/S50C) With Addition Of Nile Red
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: A1L9F |
 |
The Crystal Structure Of Cyp125Mrca, An Ancestrally Reconstructed Cyp125 Enzyme
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: LIPID BINDING PROTEIN Ligands: HEM, VD3, EDO, IPA, NA |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: TRANSPORT PROTEIN Ligands: LMT, OCT, C14, EDO, GOL, D12, HEX, XE9, D10, DDR, DDQ, SO4, LPX |
 |
Organism: Synthetic construct, Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE/RNA Ligands: ZN, GOL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: GOL, ZN |
 |
Cryoem Structure Of De Novo Antibody Fragment Scfv 6 With C. Difficile Toxin B (Tcdb)
Organism: Clostridioides difficile, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DE NOVO PROTEIN |
 |
Cryoem Structure Of De Novo Vhh, Vhh_Flu_01, Bound To Influenza Ha, Strain A/Usa:Iowa/1943 H1N1.
Organism: Synthetic construct, Influenza a virus (strain a/usa:iowa/1943 h1n1)
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DE NOVO PROTEIN Ligands: NAG |

