Search Count: 980
 |
Organism: Parasutterella secunda
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Parasutterella secunda
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG |
 |
Organism: Parasutterella secunda
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: RNA BINDING PROTEIN Ligands: MG |
 |
Cryo-Em Structure Of Brucella Abortus Lumazine Synthase (Bls) Engineered With Shiga Toxin I Subunit B (Stx1B)
Organism: Escherichia coli o157:h7, Brucella abortus biovar 1 (strain 9-941)
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: TOXIN |
 |
Cryo-Em Structure Of Brucella Abortus Lumazine Synthase (Bls) Engineered With Shiga Toxin Ii Subunit B (Stx2B)
Organism: Escherichia coli o157:h7, Brucella abortus bv. 1 str. 9-941
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: TOXIN |
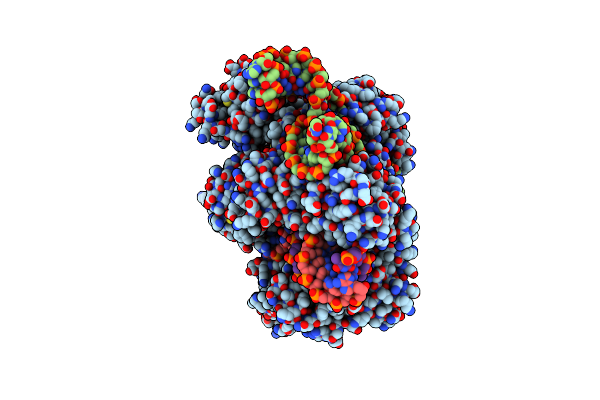 |
Organism: Parasutterella secunda, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: IMMUNE SYSTEM Ligands: MG |
 |
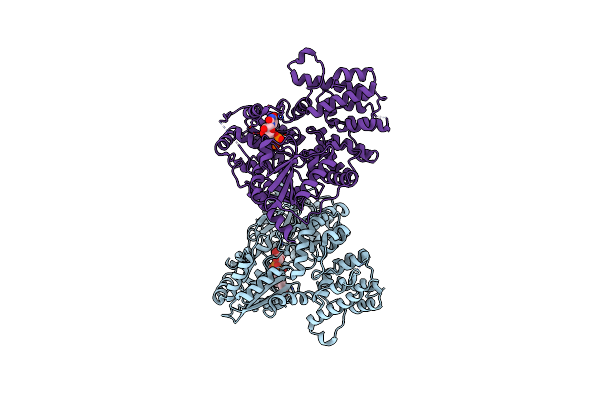
Organism: Collinsella tanakaei
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-06-12 Classification: OXIDOREDUCTASE Ligands: NAD, SO4, TRS |
 |
Crystal Structure Of Apngt With Q469A And M218A Mutations In Complex With Udp-Glc
Organism: Actinobacillus pleuropneumoniae serovar 5b str. l20
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2023-08-16 Classification: TRANSFERASE Ligands: UPG, UDP |
 |
Crystal Structure Of Powdery Mildews Blumeria Graminis F. Sp. Tritici Avrpm2(1)
Organism: Blumeria graminis f. sp. tritici
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2023-08-16 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Powdery Mildews Blumeria Graminis F. Sp. Tritici Avrpm2 (2)
Organism: Blumeria graminis f. sp. tritici
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2023-08-16 Classification: UNKNOWN FUNCTION |
 |
Organism: Brucella abortus biovar 1 (strain 9-941)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-01-12 Classification: UNKNOWN FUNCTION |
 |
Organism: Bacteroides clarus
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2021-10-06 Classification: LYASE Ligands: CA, MLI |
 |
Organism: Brucella abortus bv. 1 str. 9-941
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-06-23 Classification: HYDROLASE Ligands: H9U |
 |
Organism: Brucella abortus bv. 1 str. 9-941
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-06-23 Classification: UNKNOWN FUNCTION |
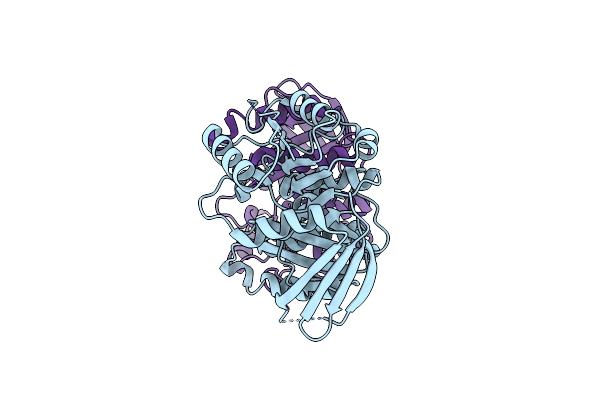 |
Crystal Structure Of Muramoyltetrapeptide Carboxypeptidase From Oxalobacter Formigenes
Organism: Oxalobacter formigenes oxcc13
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-11-04 Classification: HYDROLASE |
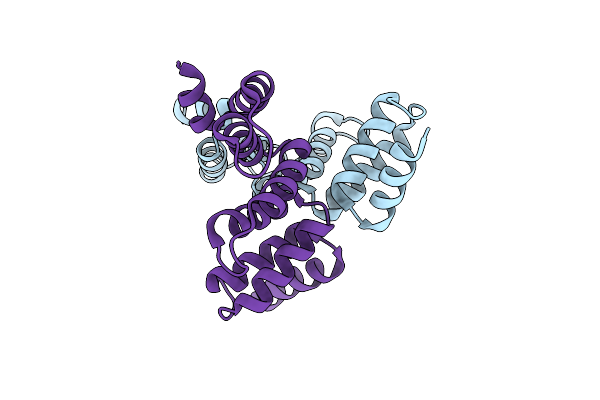 |
Organism: Oxalobacter formigenes oxcc13
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2020-05-06 Classification: UNKNOWN FUNCTION |
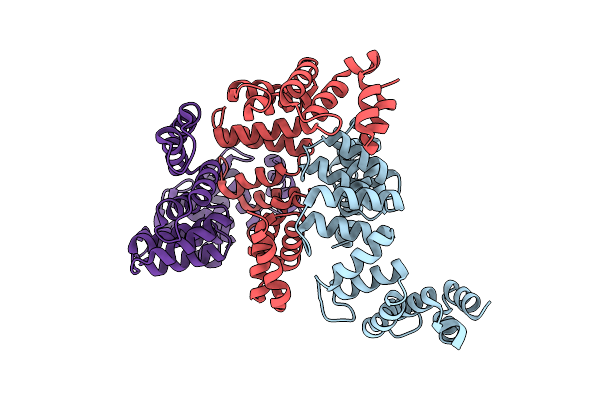 |
Organism: Oxalobacter formigenes oxcc13
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-05-06 Classification: HYDROLASE |
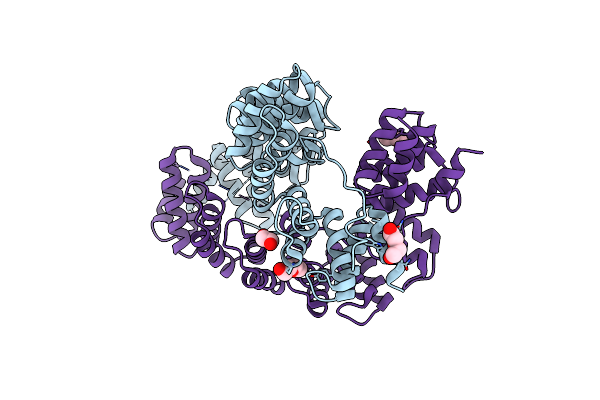 |
Organism: Oxalobacter formigenes oxcc13
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2020-04-29 Classification: HYDROLASE Ligands: PEG, EDO |
 |
Organism: Oxalobacter formigenes oxcc13
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: PGE, CL, IMD |
 |
Organism: Oxalobacter formigenes oxcc13
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: SO4, EDO, PGE, PEG |

