Search Count: 6
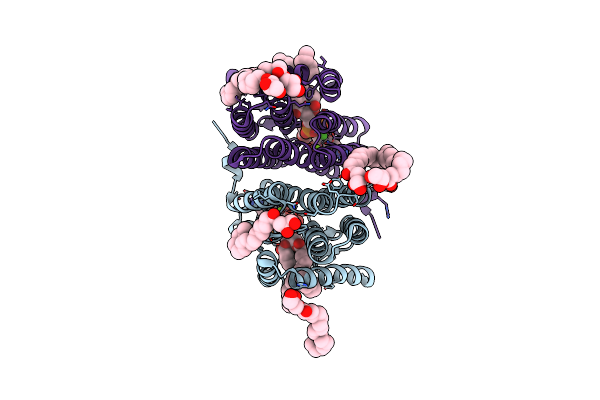 |
Crystal Structure Of Phosphatidyl Serine Synthase (Pss) In The Closed Conformation With Bound Citrate.
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Release Date: 2021-12-08 Classification: LIPID BINDING PROTEIN Ligands: 58A, CA, MG, CL, FLC, NA, OLC |
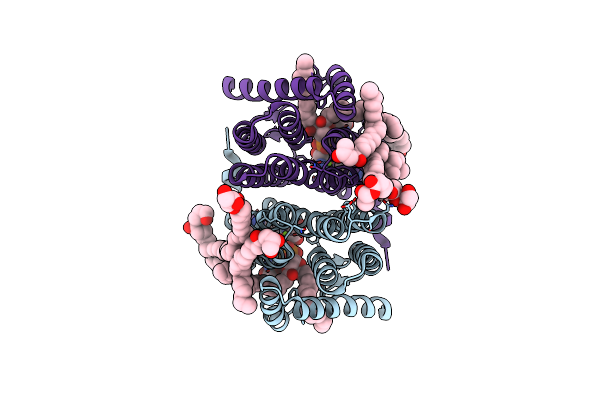 |
Crystal Structure Of Phosphatidyl Serine Synthase (Pss) In The Closed Conformation With Bound Citrate.
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-12-08 Classification: LIPID BINDING PROTEIN Ligands: 58A, CA, CL, SER, NA, MG, OLC |
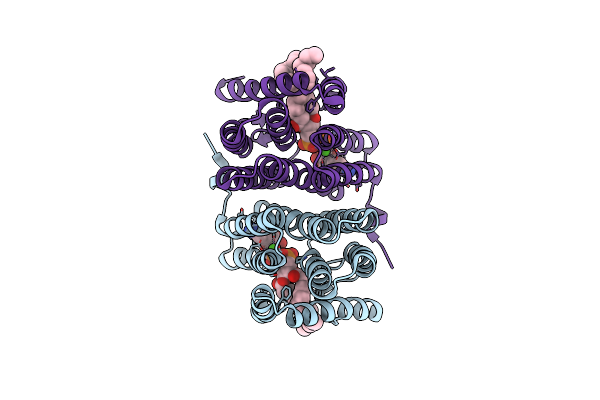 |
Crystal Structure Of Phosphatidyl Serine Synthase (Pss) In The Closed Conformation With Bound Citrate.
Organism: Methanocaldococcus jannaschii dsm 2661
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-12-08 Classification: LIPID BINDING PROTEIN Ligands: 58A, CA, CL |
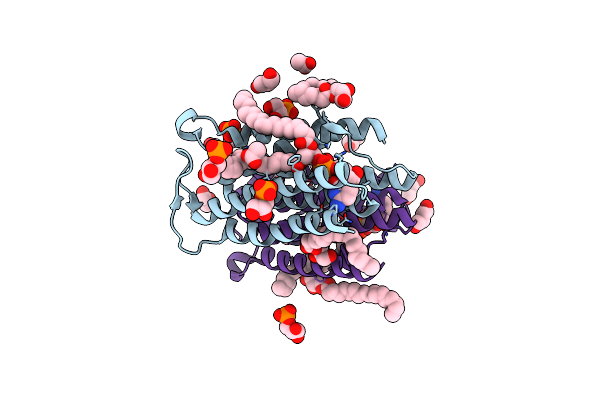 |
Crystal Structure Of Phosphatidylglycerol Phosphate Synthase In Complex With Cytidine Diphosphate-Diacylglycerol
Organism: Staphylococcus aureus (strain n315)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-12-08 Classification: TRANSFERASE Ligands: ZN, 58A, G3P, OLC, BU1, ACY |
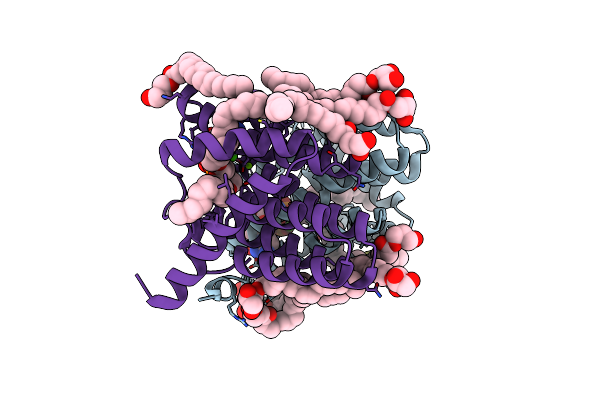 |
Crystal Structure Of Phosphatidyl Serine Synthase (Pss) In Transition State.
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2021-12-08 Classification: LIPID BINDING PROTEIN Ligands: SMW, CA, MG, CL, OLC, 58A, SER |
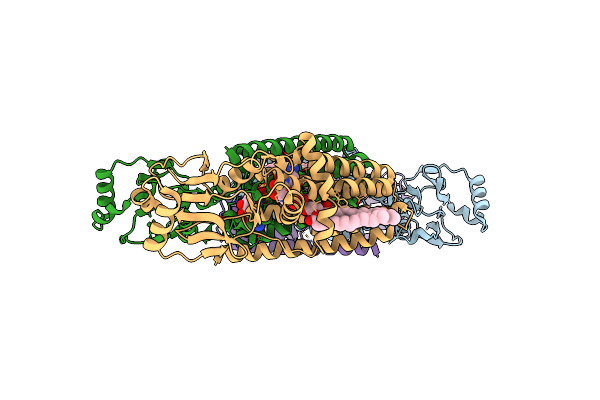 |
Structure Of A Phosphatidylinositolphosphate (Pip) Synthase From Renibacterium Salmoninarum
Organism: Archaeoglobus fulgidus, Renibacterium salmoninarum
Method: X-RAY DIFFRACTION Resolution:3.62 Å Release Date: 2015-11-04 Classification: MEMBRANE PROTEIN Ligands: 8K6, MG, 58A |

