Search Count: 6,844
 |
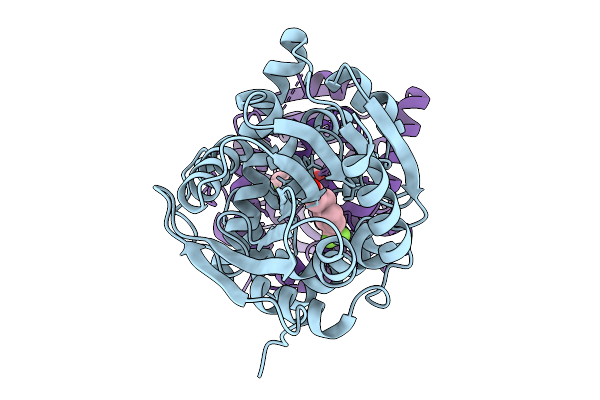
Crystal Structure Of The Fluoroacetate Dehalogenase Rpa1163 - Lys181Lys/Trp185Tyr/His280Ala With (S)-2-Fluoro-3-(4-(Trifluoromethyl)Phenyl)Propanoic Acid
Organism: Rhodopseudomonas palustris cga009
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: A1ES3 |
 |
Crystal Structure Of The Fluoroacetate Dehalogenase Rpa1163 - Lys181Ser/Trp185Gly/His280Ala With (S)-2-Fluoro-3-(4-(Trifluoromethyl)Phenyl)Propanoic Acid
Organism: Rhodopseudomonas palustris cga009
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: A1ES3 |
 |
Nmr Solution Structure Of Termini-Truncated Oxidized Cytochrome C552 From Thioalkalivibrio Paradoxus
Organism: Thioalkalivibrio paradoxus arh 1
Method: SOLUTION NMR Release Date: 2025-07-30 Classification: ELECTRON TRANSPORT Ligands: HEC |
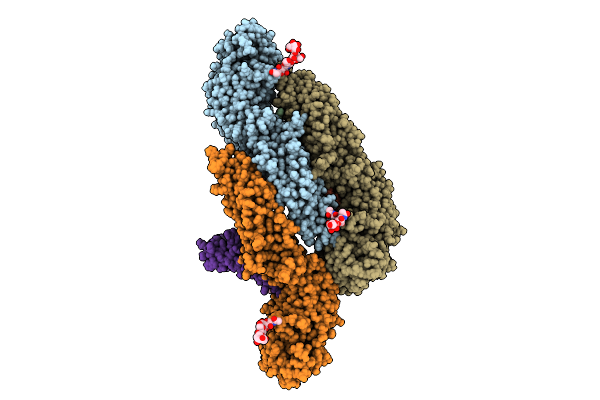 |
Organism: Deer tick virus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: VIRAL PROTEIN Ligands: CPL |
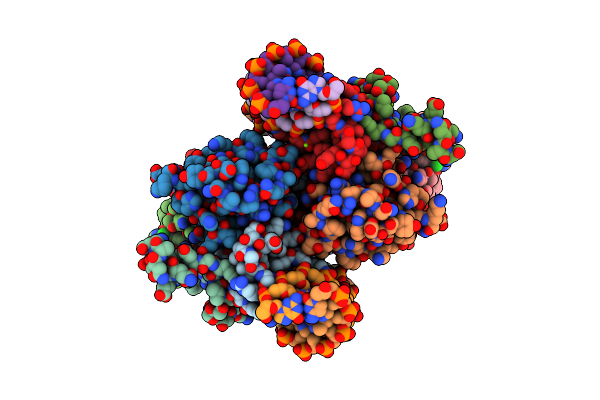 |
Organism: Proteus vulgaris
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: ANTITOXIN/DNA BINDING PROTEIN/DNA Ligands: MG, CL |
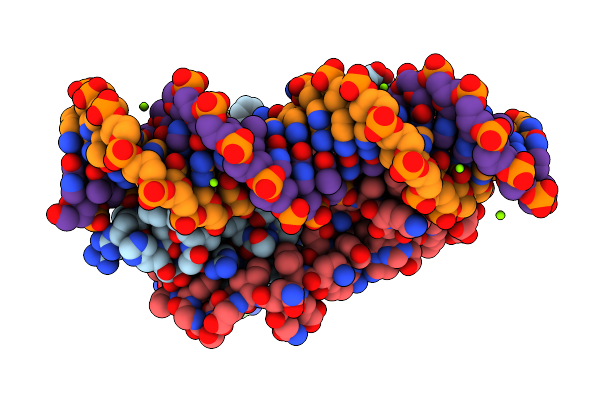 |
Organism: Proteus vulgaris
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: ANTITOXIN/DNA BINDING PROTEIN/DNA Ligands: MG |
 |
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: LIGASE Ligands: SO4, EDO |
 |
Plasmodium Vivax Aspartyl-Trna Synthetase In Complex With Aspartyl-Adenylate (Asp-Amp) Complex.
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: LIGASE Ligands: AMO, GOL, ACT |
 |
Plasmodium Vivax Aspartyl-Trna Synthetase In Complex With Aspartyl Sulfamoyl Adenosine (Asp-Ams) Complex
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: LIGASE Ligands: DSZ, MG |
 |
Cryo-Em Structure Of The Spike Glycoprotein From Bat Sars-Like Coronavirus (Bat Sl-Cov) Wiv1 In Locked State
Organism: Bat sars-like coronavirus wiv1
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: VIRAL PROTEIN Ligands: EIC, NAG |
 |
Proteus Vulgaris Tryptophan Indole-Lyase Complexed With L-Ethionine And Na+
Organism: Proteus vulgaris
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2025-04-16 Classification: LYASE Ligands: DMS, EDO, ESC, NA, YAR, YAE |
 |
Organism: Orthotospovirus tomatomaculae
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: VIRAL PROTEIN Ligands: RTP |
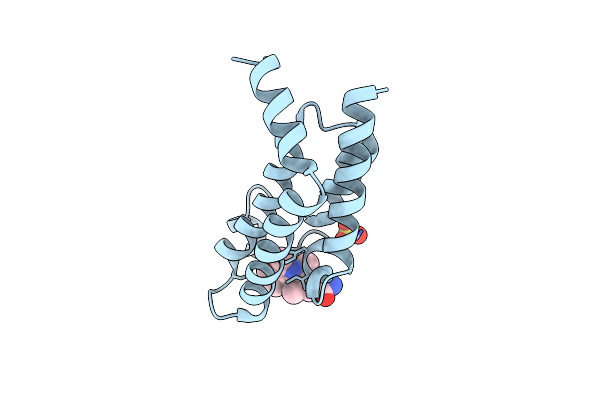 |
Crystal Structure Of The Plasmodium Vivax Bromodomain Pvbdp1 In Complex With Rmm25
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSCRIPTION Ligands: A1IUH, SO4, CL |
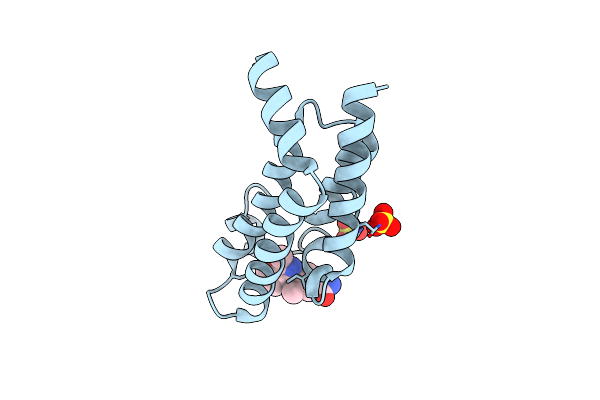 |
Crystal Structure Of The Plasmodium Vivax Bromodomain Pvbdp1 In Complex With Rmm23
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSCRIPTION Ligands: SO4, CL, A1IUW |
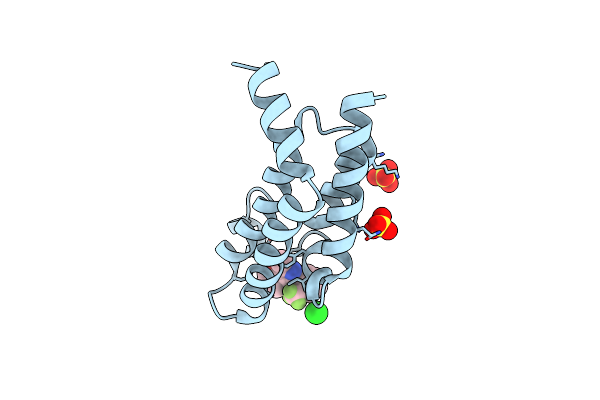 |
Crystal Structure Of The Plasmodium Vivax Bromodomain Pvbdp1 In Complex With Rmm4
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSCRIPTION Ligands: SO4, A1IUX, CL |
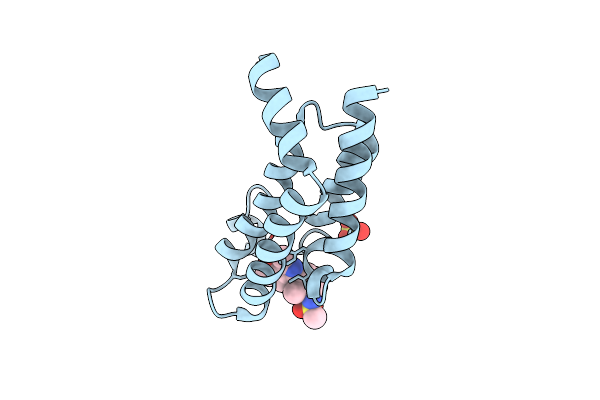 |
Crystal Structure Of The Plasmodium Vivax Bromodomain Pvbdp1 In Complex With Rmm21
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSCRIPTION Ligands: SO4, A1IUY |
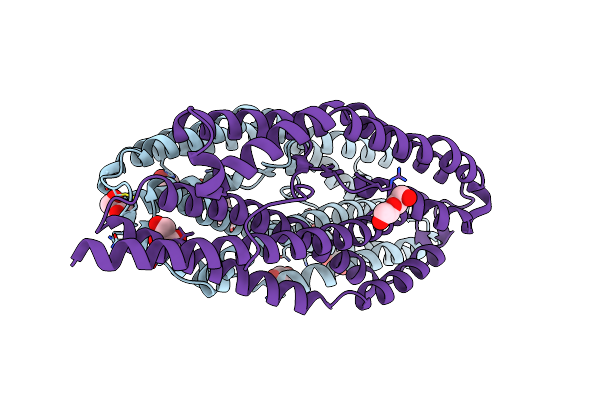 |
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-02-26 Classification: CELL ADHESION Ligands: MPD, GOL, EPE, PEG |
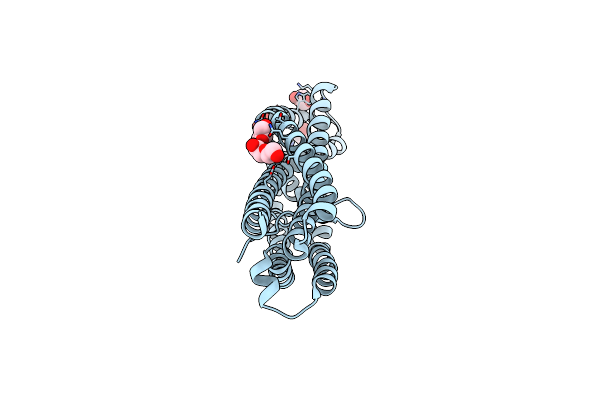 |
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-02-26 Classification: CELL ADHESION Ligands: GOL, PEG |
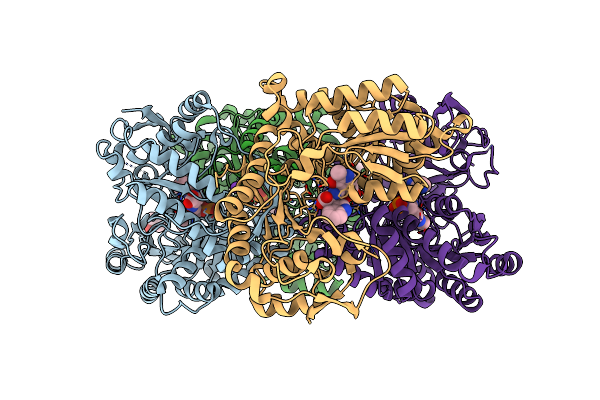 |
Proteus Vulgaris Tryptophan Indole-Lyase Complexed With L-Trp And Benzimidazole
Organism: Proteus vulgaris
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2025-02-12 Classification: LYASE Ligands: K, BZI, 0JO, DMS, PLT, A1A2Z |
 |
Organism: Plasmodium vivax, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-02-05 Classification: CELL INVASION Ligands: CA, NAG, SO4 |

