Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Morganella morganii
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: BIOSYNTHETIC PROTEIN Ligands: MPD, PEG, SO4 |
 |
Organism: Morganella morganii
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: BIOSYNTHETIC PROTEIN Ligands: PO4, PEG |
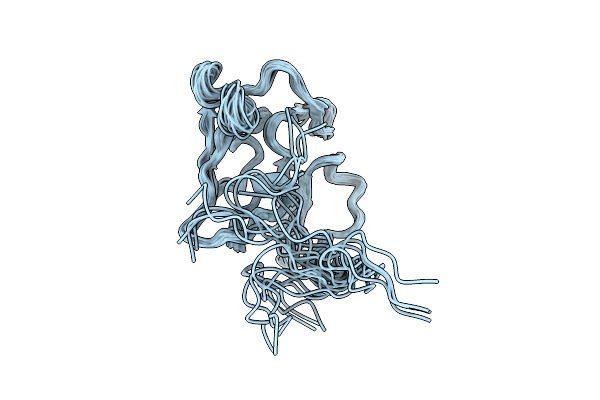 |
Three-Dimensional Structure Of The Merozoite Surface Protein 1 C-Terminal Domain
Organism: Plasmodium berghei
Method: SOLUTION NMR Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN |
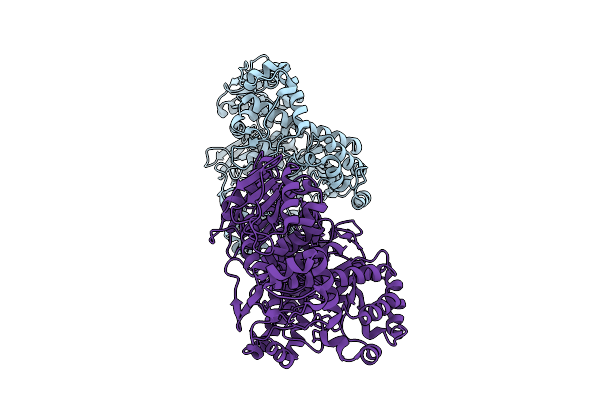 |
Organism: Wenyingzhuangia aestuarii
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: HYDROLASE |
 |
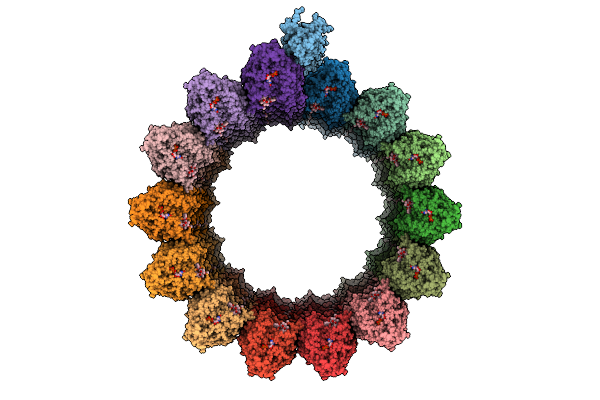
Organism: Clostridium phage phicd111
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: ANTIBIOTIC Ligands: ZN |
 |
Organism: Homo sapiens, Recombinant vesicular stomatitis indiana virus rvsv-g/gfp, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: PROTEIN BINDING Ligands: GTP, MG, GDP, TA1 |
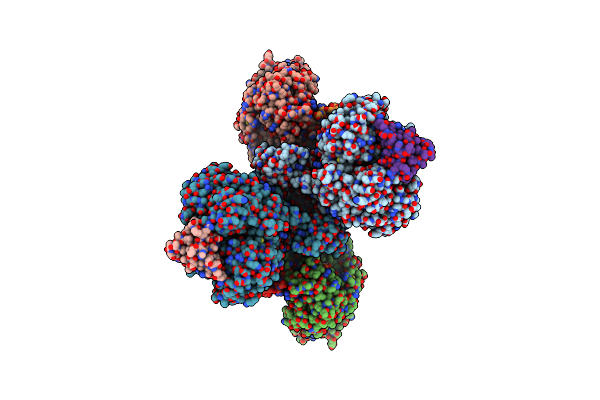 |
Cryo-Em Structure Of Reduced Form Of Formate Dehydrogenase From Rhodobacter Aestuarii (Rafdh) With Nadh
Organism: Rhodobacter aestuarii
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: OXIDOREDUCTASE Ligands: MGD, 6MO, FES, SF4, NAI, FMN |
 |
Cryo-Em Structure Of Formate Dehydrogenase From Rhodobacter Aestuarii (Rafdh) With Nad+
Organism: Rhodobacter aestuarii
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: OXIDOREDUCTASE Ligands: 2MD, MGD, 6MO, FES, SF4, NAD, FMN |
 |
Organism: Kalmanozyma brasiliensis ghg001
Method: X-RAY DIFFRACTION Resolution:3.46 Å Release Date: 2025-04-02 Classification: HYDROLASE Ligands: NAG |
 |
Vanillyl-Alcohol Dehydrogenase From Marinicaulis Flavus: P151L Mutant Bound To Eugenol
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, NO3, PEG, 1PE, PE4, EOL |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, GOL |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, PEG |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, CL |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, NO3, PEG, SO4 |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, CL, PEG |
 |
Organism: Nocardioides sp. s-1144
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-12-04 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Lacticaseibacillus casei
Method: X-RAY DIFFRACTION Release Date: 2024-11-20 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
 |
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-10-02 Classification: HYDROLASE Ligands: CA, CL, SO4, XHR |