Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
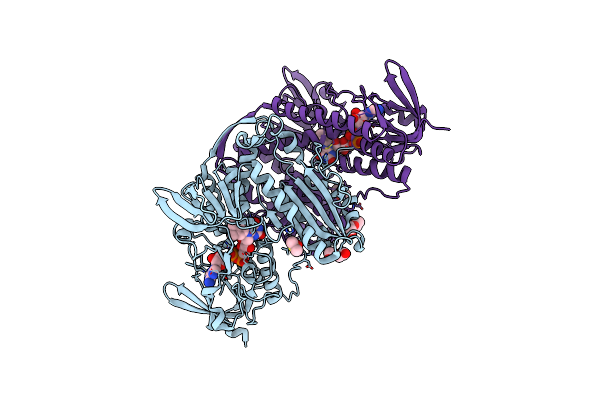 |
Crystal Structure Of Thioredoxin Reductase From Cryptosporidium Parvum In The "In" Conformation
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-05-07 Classification: FLAVOPROTEIN Ligands: FAD, DMS, PEG |
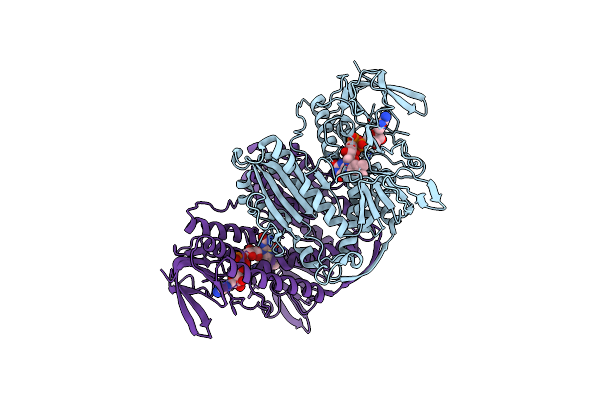 |
Crystal Structure Of Thioredoxin Reductase From Cryptosporidium Parvum In The "Waiting" Position
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2025-05-07 Classification: FLAVOPROTEIN Ligands: FAD |
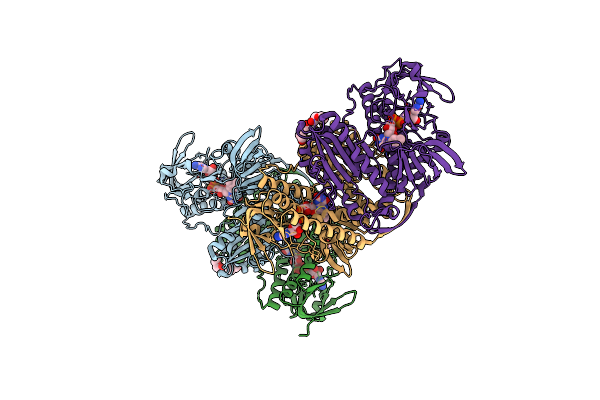 |
Crystal Structure Of Thioredoxin Reductase From Cryptosporidium Parvum In The "Activated In" Conformation
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-05-07 Classification: FLAVOPROTEIN Ligands: FAD, PEG |
 |
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Release Date: 2024-10-30 Classification: LIGASE Ligands: LYS, XLQ, GOL, DMS, SO4 |
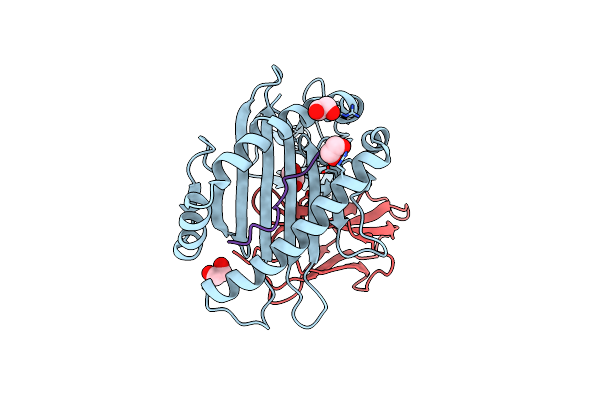 |
The First Crystal Structure Of A H-2Kb-Restricted Decapeptide From Cryptosporidium Parvum
Organism: Mus musculus, Cryptosporidium parvum
Method: X-RAY DIFFRACTION Release Date: 2024-05-29 Classification: IMMUNE SYSTEM Ligands: EDO |
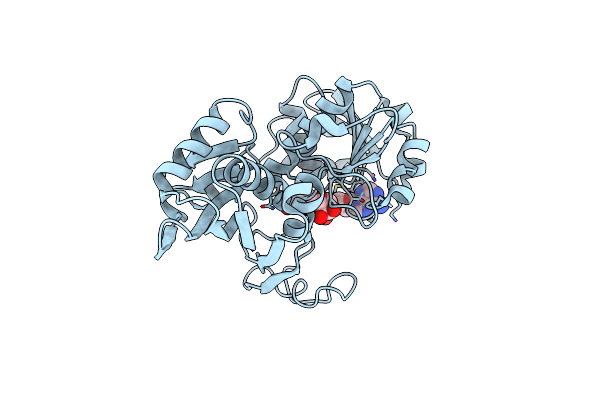 |
Organism: Cryptosporidium parvum
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2018-04-18 Classification: TRANSFERASE Ligands: R78, GOL, UNX, SO4 |
 |
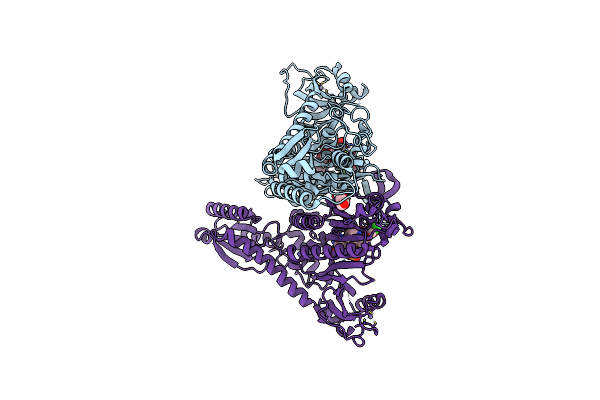
Crystal Structure Of Prolyl-Trna Synthetase From Cryptosporidium Parvum Complexed With L-Proline And Amp
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-11-16 Classification: LIGASE Ligands: PRO, AMP, ZN, PO4 |
 |
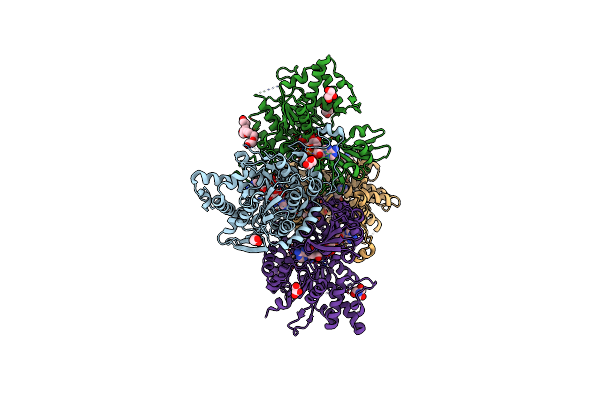
Crystal Structure Of Prolyl-Trna Synthetase From Cryptosporidium Parvum Complexed With Halofuginone And Amppnp
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-11-16 Classification: LIGASE Ligands: MG, ANP, HFG, ZN, GOL |
 |
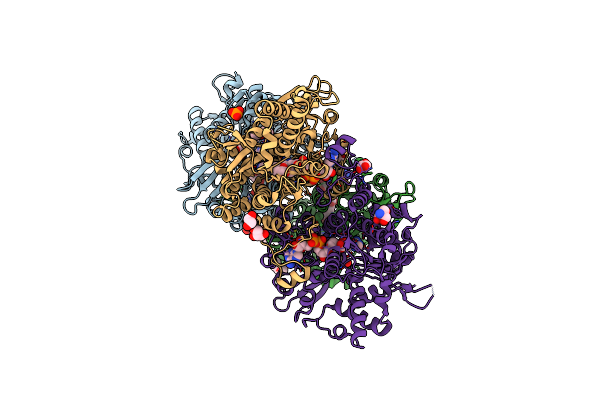
2.25 Angstrom Crystal Structure Of S-Adenosylhomocysteinase From Cryptosporidium Parvum In Complex With Neplanocin-A And Nad
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2016-05-25 Classification: HYDROLASE Ligands: 6OS, NAD, GOL, P6G, TRS |
 |
2.15 Angstrom Crystal Structure Of S-Adenosylhomocysteinase From Cryptosporidium Parvum In Complex With D-Eritadenine And Nad
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-05-18 Classification: HYDROLASE Ligands: NAD, DEA, GOL, PEG, CL, PO4, EDO, TRS |
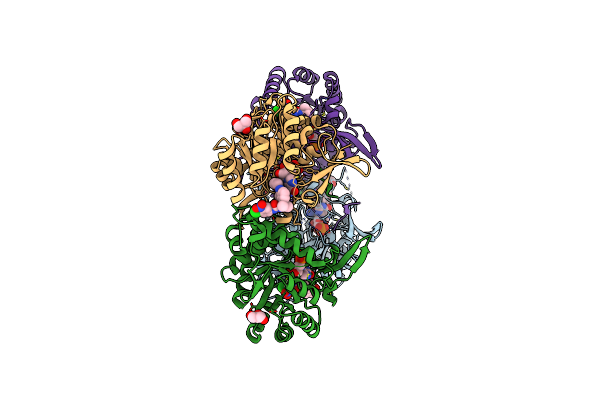 |
Co-Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Cryptosporidium Parvum And The Inhibitor P131
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2014-12-31 Classification: OXIDOREDUCTASE Ligands: IMP, I13, GOL, PEG |
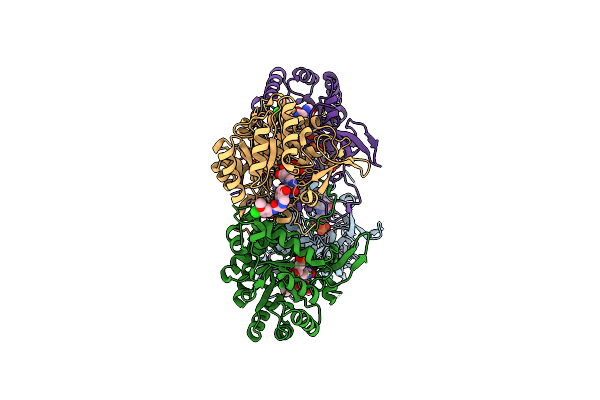 |
Co-Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Cryptosporidium Parvum With Inhibitor N109
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2014-08-06 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: IMP, N09, ACY, EDO, FMT |
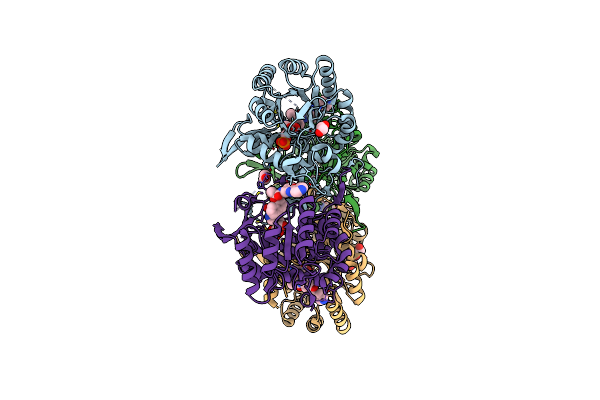 |
Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Cryptosporidium Parvum
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2013-04-03 Classification: OXIDOREDUCTASE Ligands: IMP, EDO, Q21 |
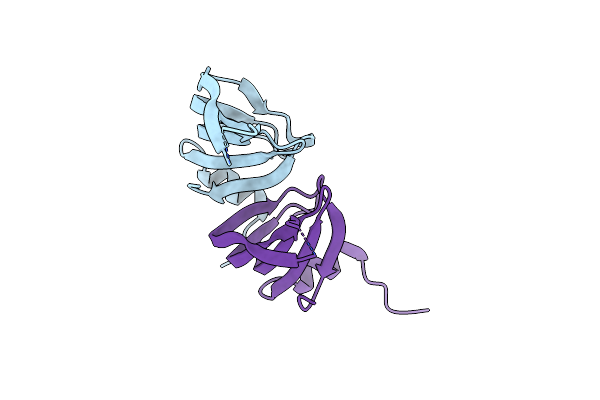 |
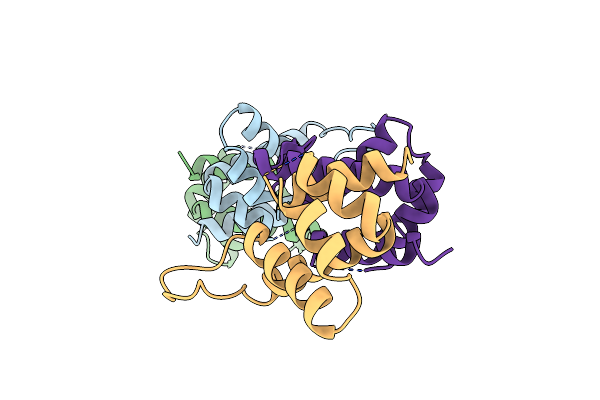
Crystal Structure Of Cryptosporidium Parvum U6 Snrna-Associated Sm-Like Protein Lsm5
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2011-02-02 Classification: DNA BINDING PROTEIN |
 |
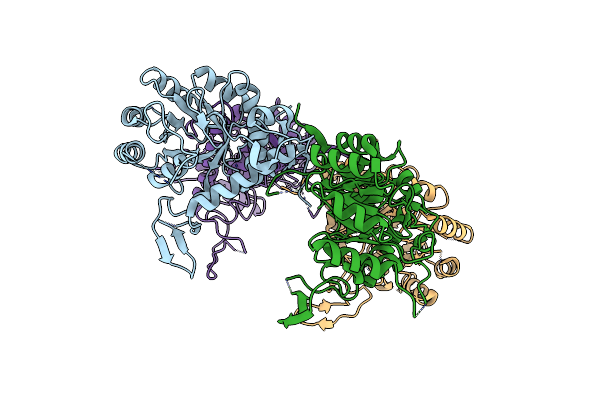
Novel Rna-Binding Domain In Cryptosporidium Parvum At 2.5 Angstrom Resolution
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2009-12-29 Classification: STRUCTURAL PROTEIN |
 |
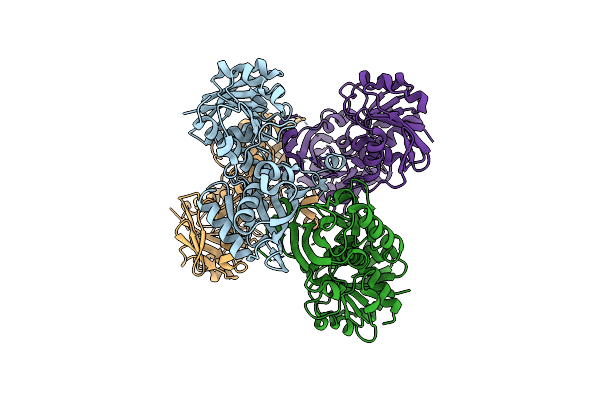
The Crystal Structure Of Cryptosporidium Parvum Inosine-5'-Monophosphate Dehydrogenase
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2009-12-15 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Apo-Glyceraldehyde 3-Phosphate Dehydrogenase From Cryptosporidium Parvum
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-03-24 Classification: OXIDOREDUCTASE |
 |
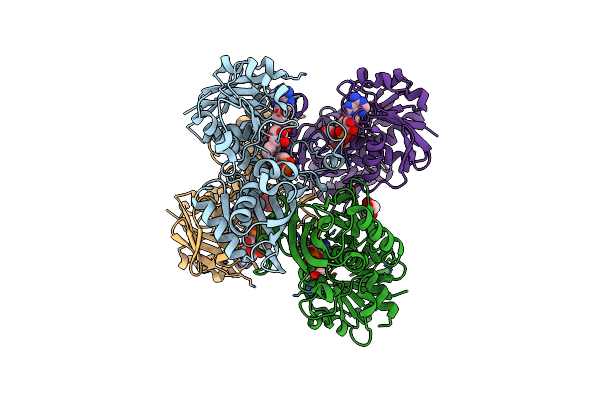
Crystal Structure Of Holo-Glyceraldehyde 3-Phosphate Dehydrogenase From Cryptosporidium Parvum
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-03-24 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Crystal Structure Of C153S Mutant Glyceraldehyde 3-Phosphate Dehydrogenase From Cryptosporidium Parvum
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-03-24 Classification: OXIDOREDUCTASE Ligands: G3H, NAD, GOL, MG |