Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
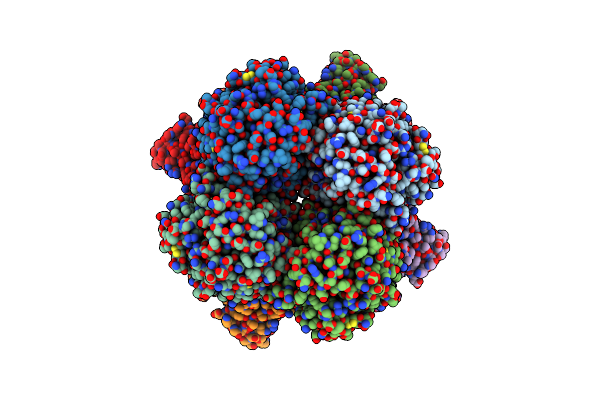
Crystal Structure Of The Coiled-Coil Based Fibril Core Of The Tardigrade Cahs-8 Protein
Organism: Hypsibius exemplaris
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: PROTEIN FIBRIL |
 |
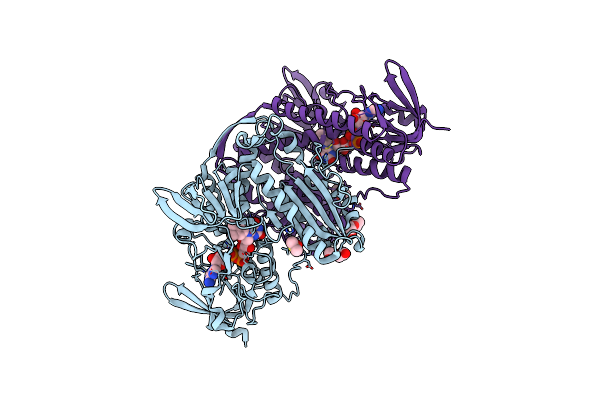
Neuraminidase Of A/Red Knot/Delaware Bay/310/2016 H10N4 In Complex With Cav-F6 Fab
Organism: Influenza a virus (a/red knot/delaware bay/310/2016(h10n4)), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, CA |
 |
Crystal Structure Of C1 Domain From Surface Protein Slpm Of Lactobacillus Brevis
Organism: Levilactobacillus brevis
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: STRUCTURAL PROTEIN |
 |
 |
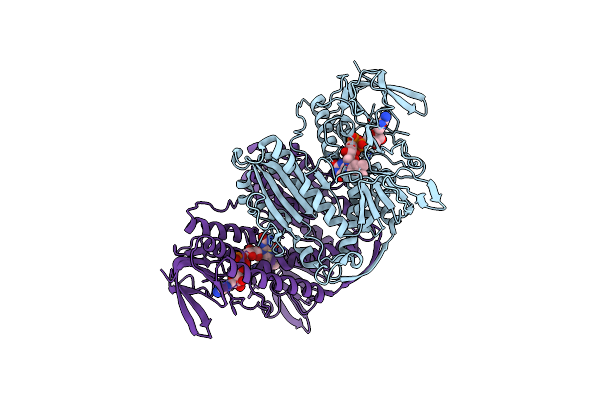
Crystal Structure Of Thioredoxin Reductase From Cryptosporidium Parvum In The "In" Conformation
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-05-07 Classification: FLAVOPROTEIN Ligands: FAD, DMS, PEG |
 |
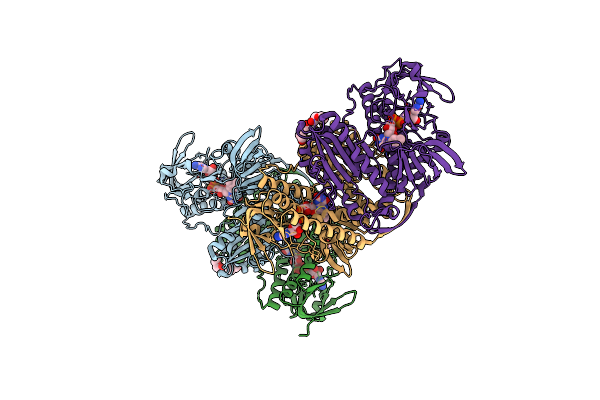
Crystal Structure Of Thioredoxin Reductase From Cryptosporidium Parvum In The "Waiting" Position
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2025-05-07 Classification: FLAVOPROTEIN Ligands: FAD |
 |
Crystal Structure Of Thioredoxin Reductase From Cryptosporidium Parvum In The "Activated In" Conformation
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-05-07 Classification: FLAVOPROTEIN Ligands: FAD, PEG |
 |
Organism: Levilactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: NAP, EDO, PEG, PGE, ACY, MG |
 |
Organism: Levilactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: NAP, SO4, MG |
 |
Organism: Levilactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: NAP, PG4, MG, EDO |
 |
Cryo-Em Structure Of Yeast Cmg Helicase Stalled At G4-Containing Dna Template, State 1
Organism: Saccharomyces cerevisiae w303, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: REPLICATION/DNA Ligands: ADP, MG, ZN, ATP |
 |
Cryo-Em Structure Of Yeast Cmg Helicase Stalled At G4-Containing Dna Template, State 3
Organism: Saccharomyces cerevisiae w303, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: REPLICATION/DNA Ligands: ADP, MG, ZN, ATP |
 |
Cryo-Em Structure Of Yeast Cmg Helicase Stalled At G4-Containing Dna Template, State 2
Organism: Saccharomyces cerevisiae w303, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: REPLICATION/DNA Ligands: ATP, MG, ZN, ADP |
 |
Organism: Homo sapiens, Human papillomavirus 11
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
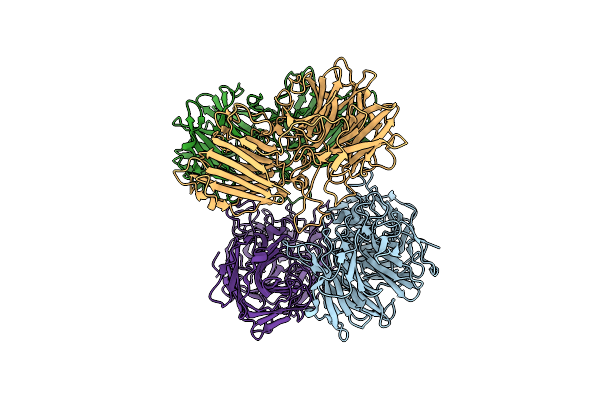 |
The Molecular Structure Of A Beta-1,4-D-Xylosidase From The Probiotic Bacterium Levilactobacillus Brevis
Organism: Levilactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-01-15 Classification: CARBOHYDRATE |
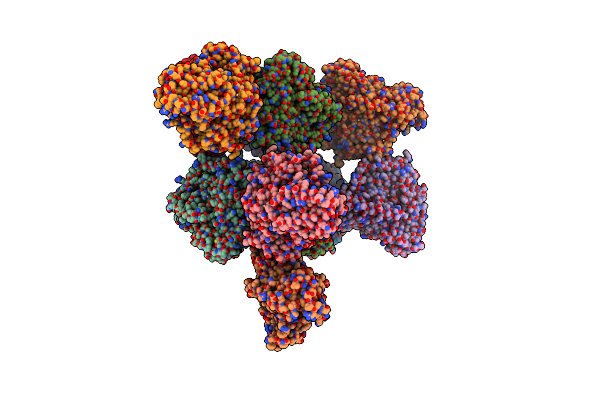 |
Organism: Enhygromyxa salina
Method: X-RAY DIFFRACTION Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: CD |
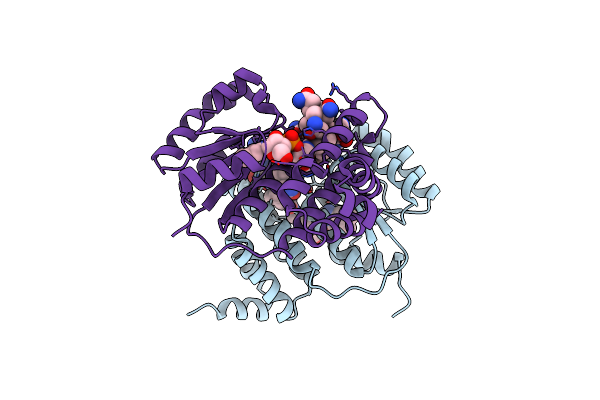 |
B12-Binding Domain From Chloracidobacterium Thermophilum Merr Family Protein, Dark State
Organism: Chloracidobacterium thermophilum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-11-13 Classification: TRANSCRIPTION Ligands: B12, 5AD |
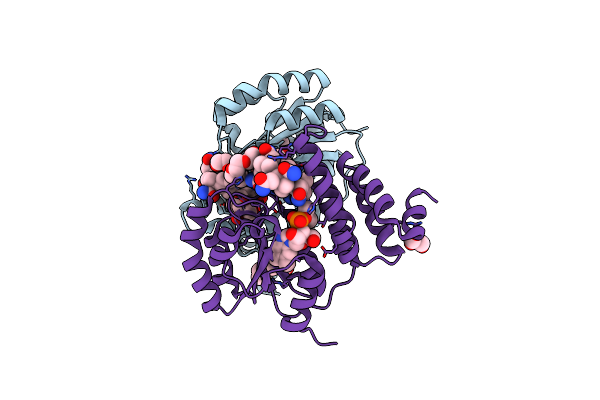 |
B12-Binding Domain From Chloracidobacterium Thermophilum Merr Family Protein, Anaerobic Light State
Organism: Chloracidobacterium thermophilum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-11-13 Classification: TRANSCRIPTION Ligands: B12, TRS, P6G, PG4, GOL |
 |
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Release Date: 2024-10-30 Classification: LIGASE Ligands: LYS, XLQ, GOL, DMS, SO4 |
 |
The Crystal Structure Of D-Mandelate Dehydrogenase From Lactobacillus Brevis
Organism: Levilactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-10-02 Classification: OXIDOREDUCTASE |