Search Count: 5,395
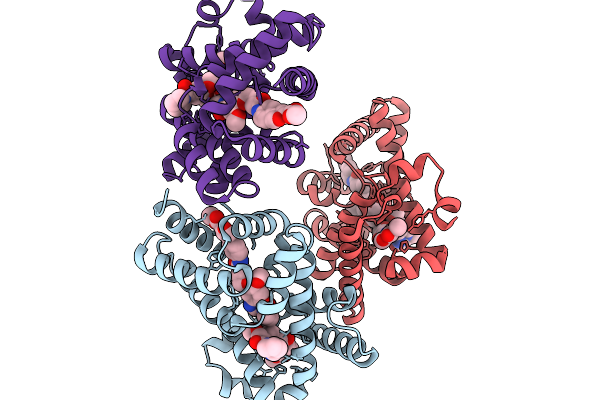 |
X-Ray Structure Of The Drug Binding Domain Of Alba In Complex With The Kmr-04-154 Compound Of The Pyrrolobenzodiazepines Class
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2026-02-04 Classification: ANTIMICROBIAL PROTEIN Ligands: A1I1F, A1IZN |
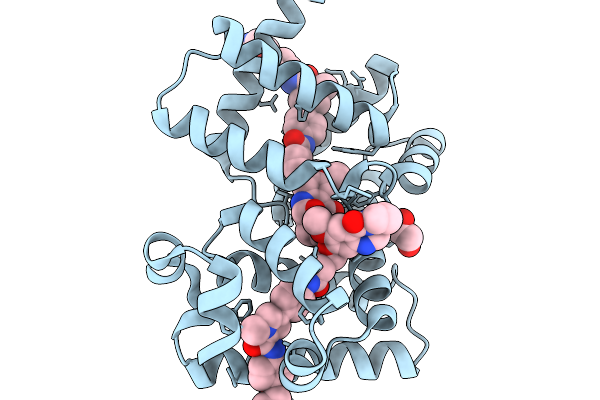 |
X-Ray Structure Of The Drug Binding Domain Of Alba In Complex With The Kmr-04-161 Compound Of The Pyrrolobenzodiazepines Class
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2026-02-04 Classification: ANTIMICROBIAL PROTEIN Ligands: A1I0A, A1I0B, EDO |
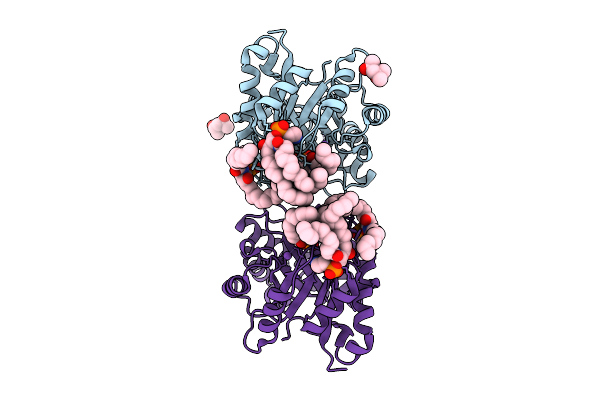 |
Structure Of Phospholipase D Betaib1I From Sicarius Terrosus Venom, H47N Mutant Bound To Product And Substrate Sphingolipids At 2.2 A Resolution From A 2-Day Old Crystal
Organism: Sicarius terrosus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-12-03 Classification: LYASE Ligands: A1A43, A1A44, NA, MG, MPD |
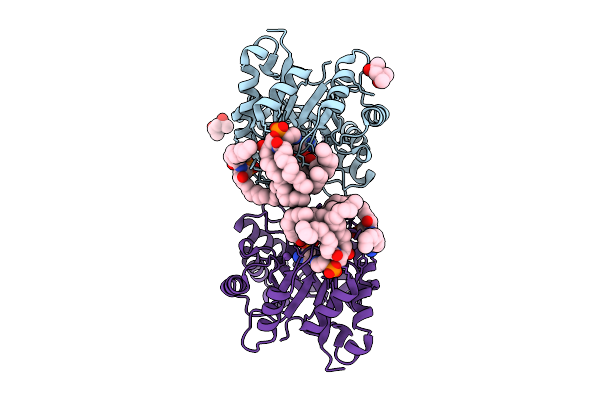 |
Structure Of Phospholipase D Betaib1I From Sicarius Terrosus Venom, H47N Mutant Bound To Substrate Sphingolipids At 2.60 A Resolution
Organism: Sicarius terrosus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-12-03 Classification: LYASE Ligands: A1A43, NA, MG, MPD |
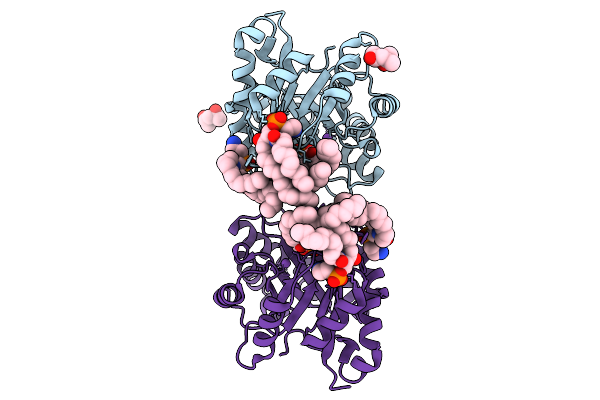 |
Structure Of Phospholipase D Betaib1I From Sicarius Terrosus Venom, H47N Mutant Bound To Product And Substrate Sphingolipids At 1.85 A Resolution
Organism: Sicarius terrosus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-09-10 Classification: LYASE Ligands: A1A43, A1A44, NA, MG, MPD |
 |
Cryoem Structure Of Calcineurin-Fusion Human Endothelin Receptor Type-B In Complex With Res-701-3
Organism: Homo sapiens, Streptomyces venezuelae
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: MEMBRANE PROTEIN Ligands: CA, FE, ZN, FK5 |
 |
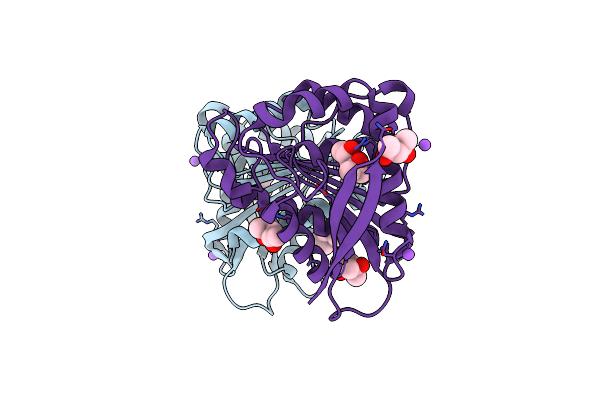
X-Ray Structure Of The Drug Binding Domain Of Alba In Complex With The Kmr-14-14 Compound Of The Pyrrolobenzodiazepines Class
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2025-01-15 Classification: DNA BINDING PROTEIN Ligands: A1H1R, A1H1Q, DTV |
 |
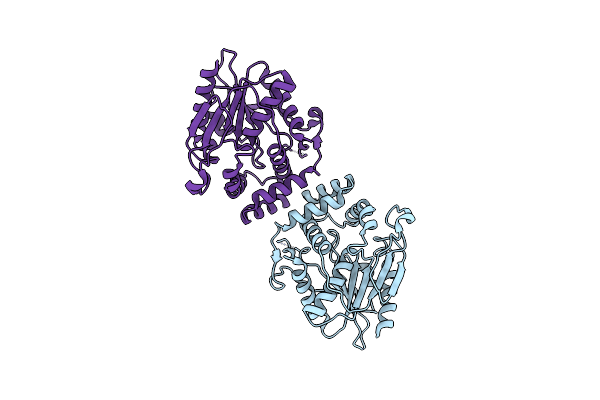
Organism: Solimonas fluminis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2024-12-18 Classification: HYDROLASE Ligands: MPD, NA |
 |
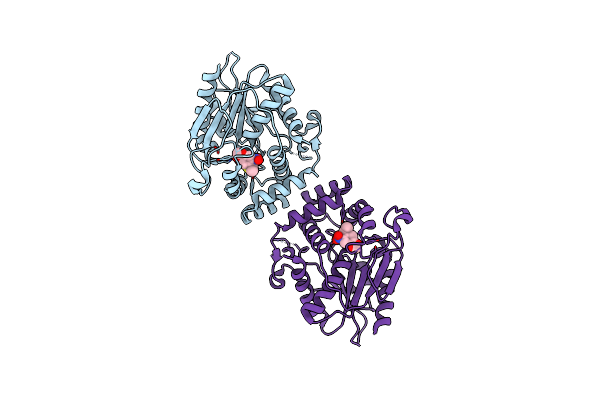
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-09-18 Classification: HYDROLASE |
 |
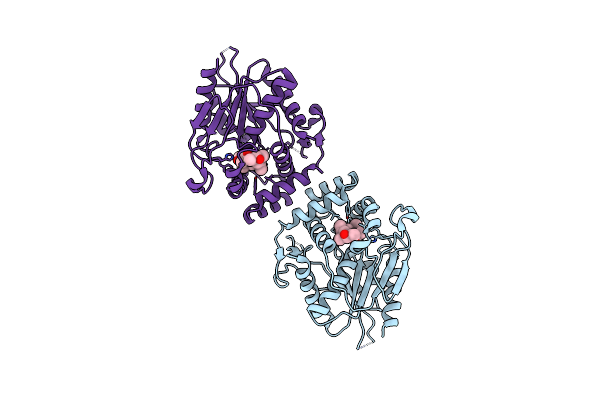
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2024-09-18 Classification: HYDROLASE Ligands: A1AWE |
 |
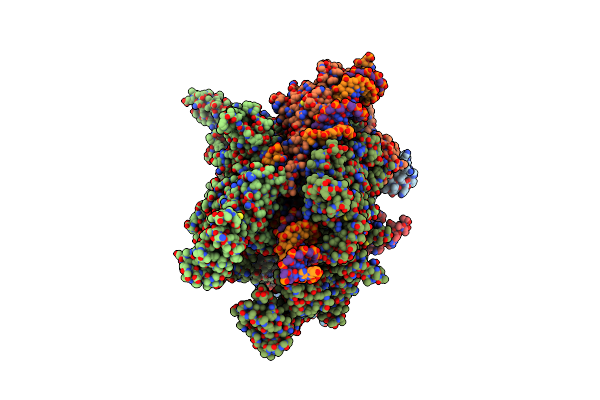
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-09-18 Classification: HYDROLASE Ligands: A1AWF |
 |
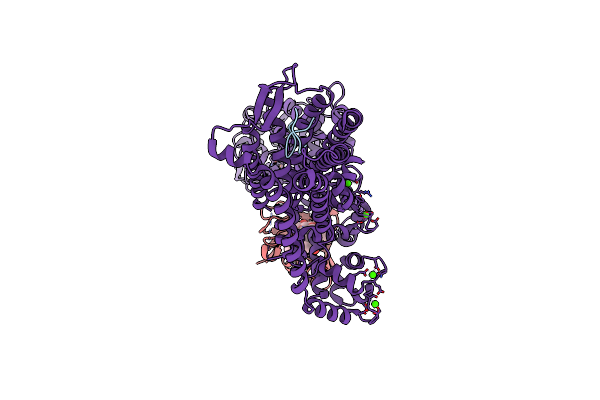
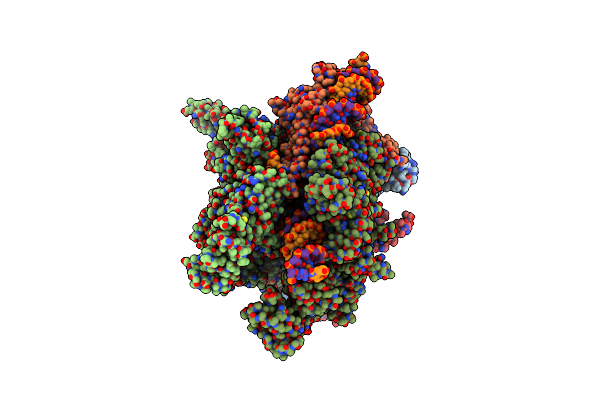
Crystal Structure Of Cis-Epoxysuccinate Hydrolase From Klebsiella Oxytoca With L(+)-Tartaric Acid
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2024-06-12 Classification: HYDROLASE Ligands: TLA, EDO, GOL, CO3 |
 |
Cryo-Em Structure Of Bacterial Rna Polymerase-Sigma54 Initial Transcribing Complex - 5Nt Pre-Translocated Complex
Organism: Escherichia coli k-12, Klebsiella oxytoca
Method: ELECTRON MICROSCOPY Release Date: 2024-01-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Cryo-Em Structure Of Bacterial Rna Polymerase-Sigma54 Initial Transcribing Complex - 5Nt Post-Translocated Complex
Organism: Escherichia coli k-12, Klebsiella oxytoca
Method: ELECTRON MICROSCOPY Release Date: 2024-01-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Cryo-Em Structure Of Bacterial Rna Polymerase-Sigma54 Initial Transcribing Complex - 6Nt Complex
Organism: Escherichia coli k-12, Klebsiella oxytoca
Method: ELECTRON MICROSCOPY Release Date: 2024-01-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Cryo-Em Structure Of Bacterial Rna Polymerase-Sigma54 Initial Transcribing Complex - 7Nt Complex
Organism: Klebsiella oxytoca, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2024-01-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Cryo-Em Structure Of Bacterial Rna Polymerase-Sigma54 Initial Transcribing Complex - 8Nt Complex
Organism: Klebsiella oxytoca, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2024-01-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Cryo-Em Structure Of Bacterial Rna Polymerase-Sigma54 Initial Transcribing Complex - 9Nt Complex
Organism: Escherichia coli k-12, Klebsiella oxytoca
Method: ELECTRON MICROSCOPY Release Date: 2024-01-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Crystal Structure Of 4-Amino-4-Deoxychorismate Synthase From Streptomyces Venezuelae
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: TRP, MLT, MG, SIN |
 |
Crystal Structure Of 4-Amino-4-Deoxychorismate Synthase From Streptomyces Venezuelae Co-Crystallized With L-Glutamine
Organism: Streptomyces venezuelae
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: TRP, SIN, MG, CL |

