Search Count: 155
 |
Organism: Achromobacter xylosoxidans
Method: SOLUTION NMR Release Date: 2023-10-11 Classification: ANTIBIOTIC |
 |
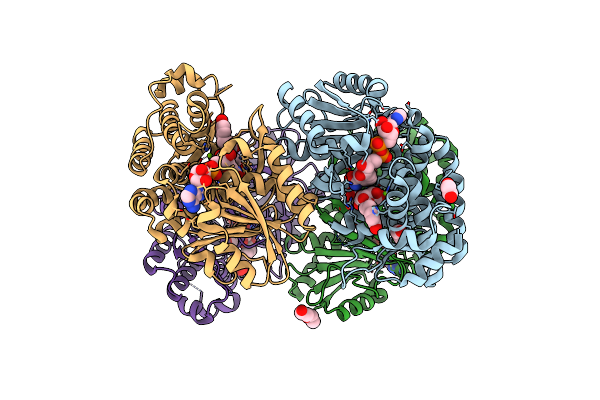
Cryoem Structure Of Quinol-Dependent Nitric Oxide Reductase (Qnor) From Alcaligenes Xylosoxidans At 2.2 A Resolution
Organism: Achromobacter xylosoxidans
Method: ELECTRON MICROSCOPY Resolution:2.20 Å Release Date: 2023-08-16 Classification: MEMBRANE PROTEIN Ligands: HEM, FE, CA, 10M, UQ1, LOP |
 |
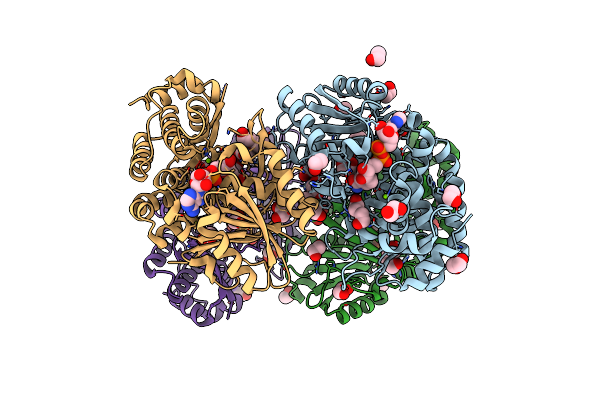
Crystal Structure Of Providencia Alcalifaciens 3-Dehydroquinate Synthase (Dhqs) In Complex With Mg2+, Nad And Chlorogenic Acid
Organism: Providencia alcalifaciens f90-2004
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-07-29 Classification: LYASE Ligands: NAD, MG, 7LH, EDO, PEG |
 |
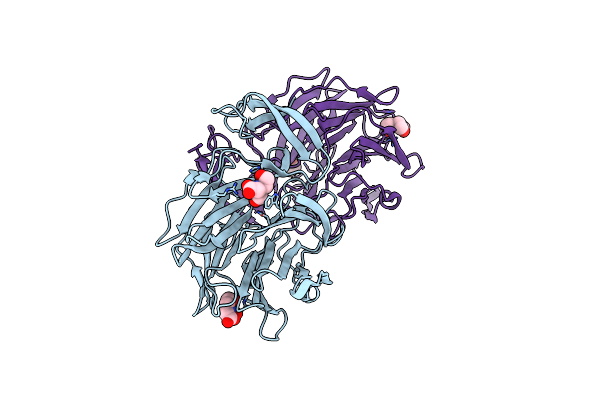
Crystal Structure Of Providencia Alcalifaciens 3-Dehydroquinate Synthase (Dhqs) In Complex With Mg2+ And Nad
Organism: Providencia alcalifaciens f90-2004
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2020-07-29 Classification: LYASE Ligands: NAD, MG, EDO, PEG |
 |
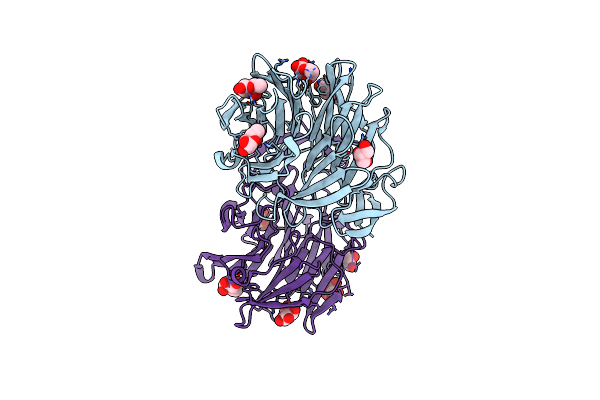
Organism: Arthrobotrys oligospora (strain atcc 24927 / cbs 115.81 / dsm 1491)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2020-07-29 Classification: SUGAR BINDING PROTEIN Ligands: BCN |
 |
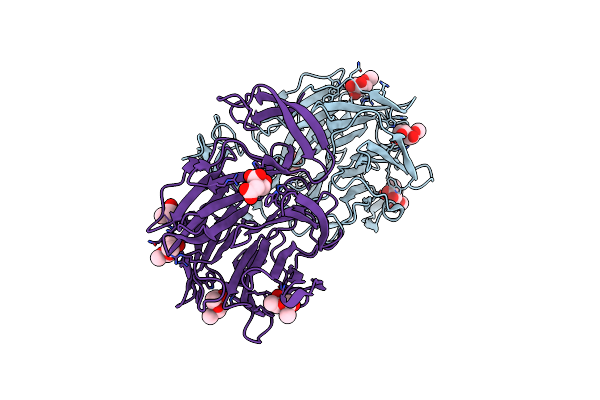
Crystal Structure Of Aoflea From Arthrobotrys Oligospora In Complex With L-Fucose
Organism: Arthrobotrys oligospora (strain atcc 24927 / cbs 115.81 / dsm 1491)
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2020-07-29 Classification: SUGAR BINDING PROTEIN Ligands: FUC, FUL, GOL |
 |
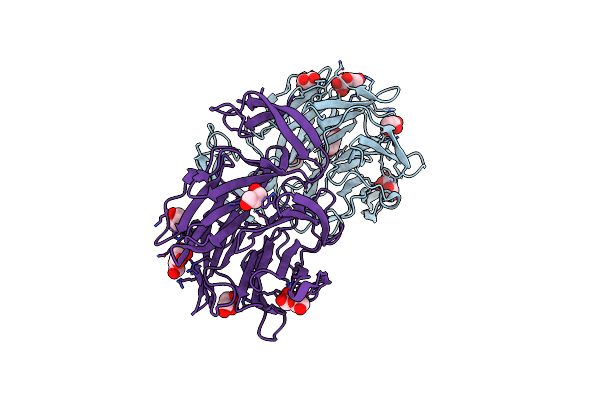
Crystal Structure Of Aoflea From Arthrobotrys Oligospora In Complex With Methylated L-Fucose
Organism: Arthrobotrys oligospora (strain atcc 24927 / cbs 115.81 / dsm 1491)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-07-29 Classification: SUGAR BINDING PROTEIN Ligands: CIT, MFU, GOL |
 |
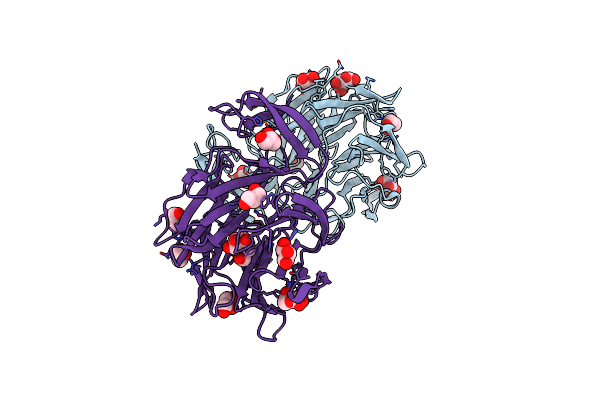
Crystal Structure Of Aoflea From Arthrobotrys Oligospora In Complex With D-Manose
Organism: Arthrobotrys oligospora (strain atcc 24927 / cbs 115.81 / dsm 1491)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-07-29 Classification: SUGAR BINDING PROTEIN Ligands: MAN, GOL, BMA |
 |
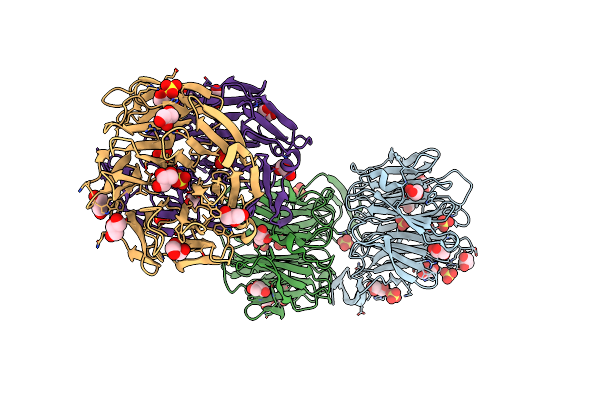
Crystal Structure Of Aoflea From Arthrobotrys Oligospora In Complex With D-Arabinose
Organism: Arthrobotrys oligospora (strain atcc 24927 / cbs 115.81 / dsm 1491)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-07-29 Classification: SUGAR BINDING PROTEIN Ligands: SEJ, 64K, GOL |
 |
Crystal Structure Of R97A/R150A/R203A Mutant Of Aoflea From Arthrobotrys Oligospora
Organism: Arthrobotrys oligospora (strain atcc 24927 / cbs 115.81 / dsm 1491)
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2020-07-29 Classification: SUGAR BINDING PROTEIN Ligands: GOL, SO4 |
 |
Glu-494-Ala Inactive Monomer Of A Quinol Dependent Nitric Oxide Reductase (Qnor) From Alcaligenes Xylosoxidans
Organism: Alcaligenes xylosoxydans xylosoxydans
Method: ELECTRON MICROSCOPY Release Date: 2020-04-01 Classification: OXIDOREDUCTASE Ligands: HEM, CA |
 |
Organism: Alcaligenes xylosoxydans xylosoxydans
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-02-05 Classification: ELECTRON TRANSPORT Ligands: CU |
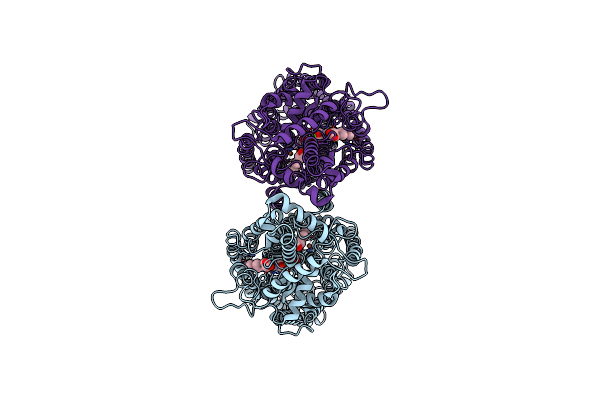 |
Cryo-Em Structure Of Dimeric Quinol Dependent Nitric Oxide Reductase (Qnor) From Alcaligenes Xylosoxidans
Organism: Alcaligenes xylosoxydans xylosoxydans
Method: ELECTRON MICROSCOPY Release Date: 2019-09-11 Classification: OXIDOREDUCTASE Ligands: HEM, FE, CA |
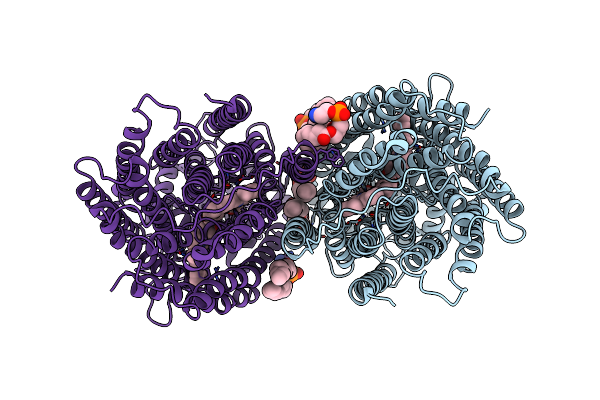 |
Cryo-Em Structure Of Dimeric Quinol Dependent Nitric Oxide Reductase (Qnor) Val495Ala Mutant From Alcaligenes Xylosoxidans
Organism: Alcaligenes xylosoxydans xylosoxydans
Method: ELECTRON MICROSCOPY Release Date: 2019-09-11 Classification: OXIDOREDUCTASE Ligands: HEM, FE, CA, LOP, LMT |
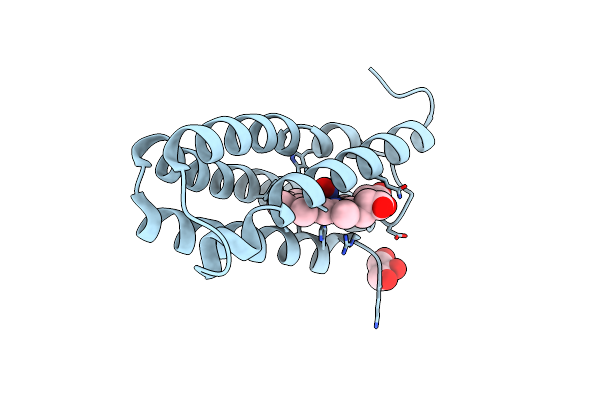 |
The 1.06 A Resolution Structure Of The L16G Mutant Of Ferric Cytochrome C Prime From Alcaligenes Xylosoxidans, Complexed With Nitrite
Organism: Alcaligenes xylosoxydans xylosoxydans
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2018-05-16 Classification: OXIDOREDUCTASE Ligands: HEC, NO2, GOL |
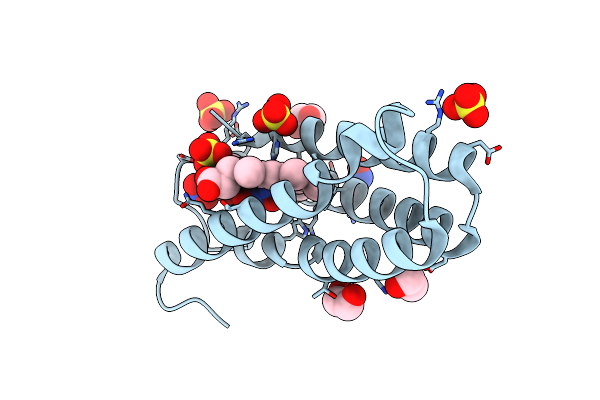 |
The 0.91 A Resolution Structure Of The L16G Mutant Of Cytochrome C Prime From Alcaligenes Xylosoxidans, Complexed With Nitric Oxide
Organism: Alcaligenes xylosoxydans xylosoxydans
Method: X-RAY DIFFRACTION Resolution:0.91 Å Release Date: 2018-01-10 Classification: OXIDOREDUCTASE Ligands: HEC, NO, NO3, NO2, SO4, EDO |
 |
Resting State Copper Nitrite Reductase Determined By Serial Femtosecond Rotation Crystallography
Organism: Alcaligenes xylosoxydans xylosoxydans
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-11-29 Classification: OXIDOREDUCTASE Ligands: CU, ZN, OXY, MES, PG4 |
 |
As-Isolated Resting State Copper Nitrite Reductase From Achromobacter Xylosoxidans
Organism: Alcaligenes xylosoxydans xylosoxydans
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-11-29 Classification: OXIDOREDUCTASE Ligands: CU, ZN, MES, PG4 |
 |
Nitric Oxide Complex Of The L16A Mutant Of Cytochrome C Prime From Alcaligenes Xylosoxidans
Organism: Alcaligenes xylosoxydans xylosoxydans
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2017-03-08 Classification: ELECTRON TRANSPORT Ligands: HEC, ASC, SO4, NO |
 |
Ferrous Leu 16 Val Mutant Of Cytochrome C Prime From Alcaligenes Xylosoxidans
Organism: Alcaligenes xylosoxydans xylosoxydans
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2017-03-08 Classification: ELECTRON TRANSPORT Ligands: HEC |

