Search Count: 74
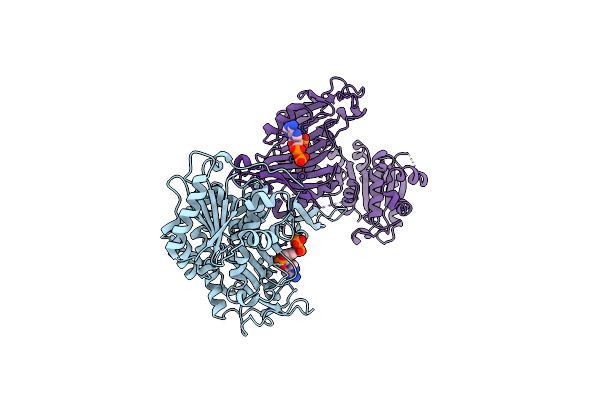 |
Glucose-6-Phosphate 1-Dehydrogenase (G6Pdh) From Crithidia Fasciculata (Nadp Bound)
Organism: Crithidia fasciculata
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-09-13 Classification: OXIDOREDUCTASE Ligands: NAP |
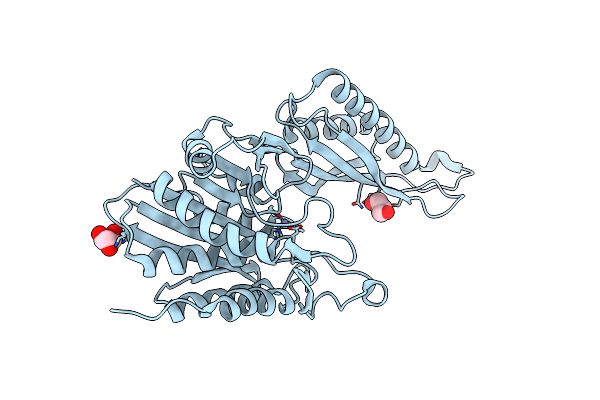 |
Organism: Enterococcus faecium 1,231,410
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-04-12 Classification: HYDROLASE Ligands: ZN, GOL |
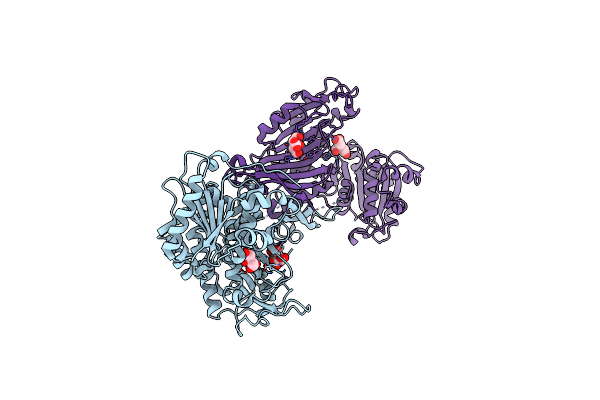 |
Glucose-6-Phosphate 1-Dehydrogenase (G6Pdh) From Crithidia Fasciculata (Citrate Bound)
Organism: Crithidia fasciculata
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2023-02-08 Classification: OXIDOREDUCTASE Ligands: CIT, CL |
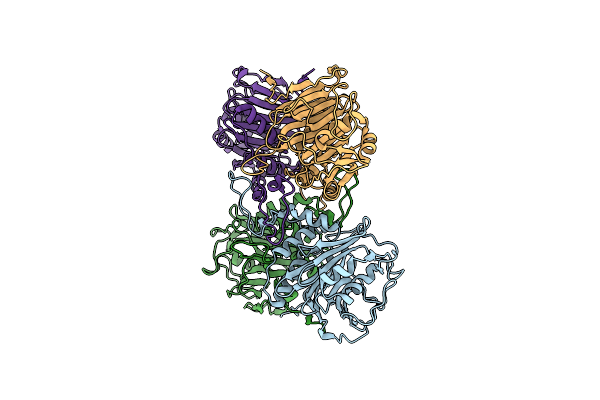 |
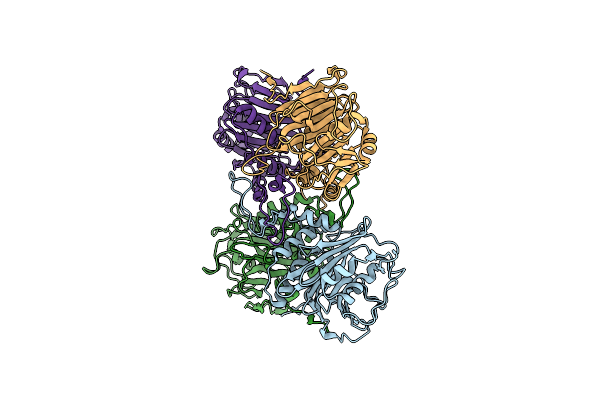
Organism: Enterococcus faecalis t2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-07-10 Classification: HYDROLASE |
 |
Organism: Enterococcus faecalis t2
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-07-10 Classification: HYDROLASE |
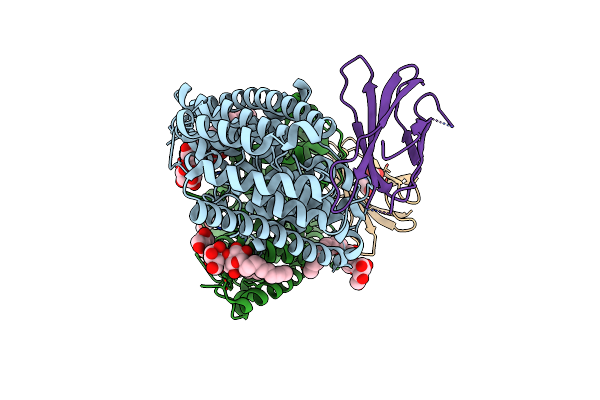 |
Organism: Enterococcus casseliflavus (strain ec10), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-07-04 Classification: TRANSPORT PROTEIN Ligands: DMU, F, MHA |
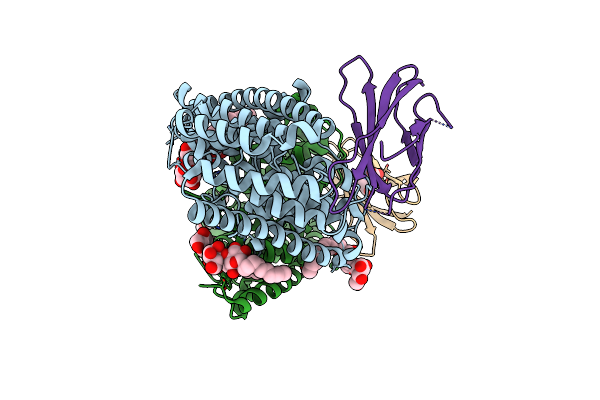 |
Organism: Enterococcus casseliflavus (strain ec10), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2018-07-04 Classification: TRANSPORT PROTEIN Ligands: DMU, F, MHA |
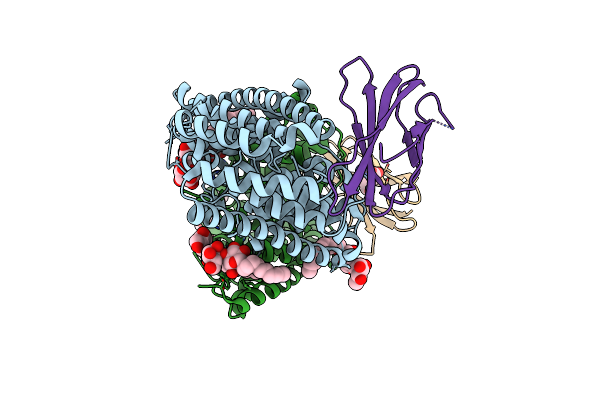 |
Organism: Enterococcus casseliflavus (strain ec10), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2018-07-04 Classification: TRANSPORT PROTEIN Ligands: DMU, F, MHA |
 |
Crystal Structure Determination Of Bile Salt Hydrolase From Enterococcus Feacalis
Organism: Enterococcus faecalis t2
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2015-11-11 Classification: HYDROLASE |
 |
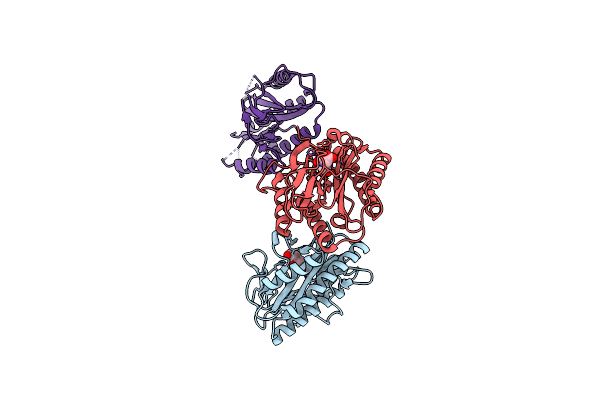
Crystal Structure Of Enolase Egbg_01401 (Target Efi-502226) From Enterococcus Gallinarum Eg2
Organism: Enterococcus gallinarum eg2
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2012-11-07 Classification: ISOMERASE Ligands: MG, GOL, CL |
 |
Crystal Structure Of The Enterococcus Faecalis Methionine Aminopeptidase Apo Form
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-08-08 Classification: HYDROLASE Ligands: CIT |
 |
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-06-22 Classification: DNA BINDING PROTEIN Ligands: CA |
 |
Organism: Crithidia fasciculata
Method: SOLUTION NMR Release Date: 2003-08-28 Classification: ELECTRON TRANSPORT |
 |
Organism: Crithidia fasciculata
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2003-08-01 Classification: OXIDOREDUCTASE |
 |
Organism: Crithidia fasciculata
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2003-04-24 Classification: ELECTRON TRANSPORT |
 |
Organism: Crithidia fasciculata
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2003-04-24 Classification: ELECTRON TRANSPORT |
 |
Organism: Crithidia fasciculata
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2003-04-24 Classification: ELECTRON TRANSPORT |
 |
Organism: Crithidia fasciculata
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2003-04-02 Classification: ELECTRON TRANSPORT Ligands: SO4 |
 |
Organism: Crithidia fasciculata
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2003-03-20 Classification: ELECTRON TRANSPORT |
 |
Organism: Crithidia fasciculata
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2002-12-19 Classification: ELECTRON TRANSPORT Ligands: SO4 |

