Search Count: 2,649
 |
Organism: Helicobacter pylori g27
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: HYDROLASE |
 |
Organism: Helicobacter pylori g27
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: HYDROLASE |
 |
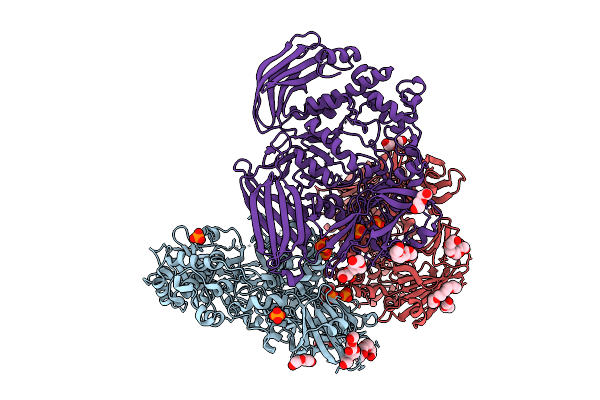
Cryo-Em Structure Of Ctpa S300A/K325A/Q329A Mutant From Helicobacter Pylori
Organism: Helicobacter pylori g27, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: HYDROLASE |
 |
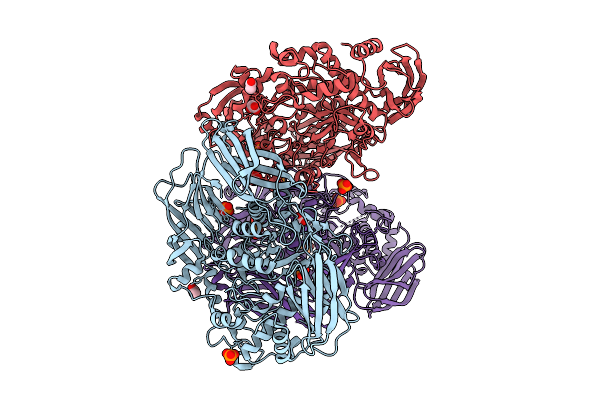
Organism: Helicobacter pylori g27
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: HYDROLASE |
 |
Organism: Helicobacter pylori g27
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: HYDROLASE |
 |
Active Conformation Of A Redox-Regulated Glycoside Hydrolase (Capgh2B) From The Gh2 Family
|
 |
Cryoem Structure Of De Novo Vhh, Vhh_Flu_01, Bound To Influenza Ha, Strain A/Usa:Iowa/1943 H1N1.
Organism: Synthetic construct, Influenza a virus (strain a/usa:iowa/1943 h1n1)
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DE NOVO PROTEIN Ligands: NAG |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group I213 At 2.05 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, GOL |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B - E553Q Mutant) From The Gh2 Family In The Space Group I213 At 2.6 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4 |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group I213 At 2.75 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, TAU |
 |
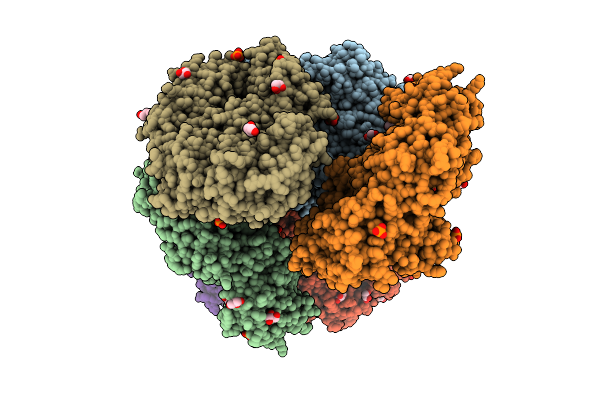
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group R3 At 2.45 A
|
 |
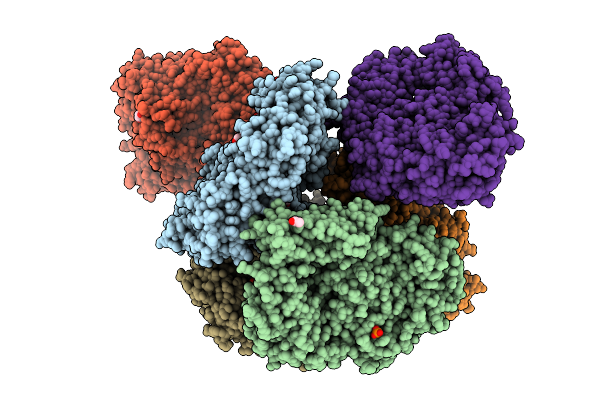
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group I212121 At 2.65 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, PEG, PG4 |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B - E553Q Mutant) From The Gh2 Family In The Space Group P3121 At 3.05 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, ACT, MLI |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group P1 At 2.40 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, GOL |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B) From The Gh2 Family In The Space Group P1 At 2.15 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, GOL, ACT |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B - E553Q Mutant) From The Gh2 Family In The Space Group P1 At 2.25 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, EDO |
 |
Crystal Structure Of The Inactive Conformation Of A Glycoside Hydrolase (Capgh2B - E465A Mutant) From The Gh2 Family In The Space Group P1 At 3.1 A
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: PO4, EDO |
 |
Neutron Structure Of Gh1 Beta-Glucosidase Td2F2 Ligand-Free Form At Room Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: NHE, DOD |
 |
Neutron Structure Of Gh1 Beta-Glucosidase Td2F2 Glucose Complex At Room Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: BGC, NHE, DOD |
 |
Neutron Structure Of Gh1 Beta-Glucosidase Td2F2 2F-Glc Complex At Room Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: G2F, NHE, DOD |

