Search Count: 4,423
 |
Organism: Natrinema sp. j7-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: PROTEIN FIBRIL |
 |
Organism: Burkholderia cenocepacia h111
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: METAL BINDING PROTEIN Ligands: FE, ACY, FMT, NA |
 |
Crystal Structure Of The Murt/Gatd Enzyme Complex From Streptococcus Pyogenes
Organism: Streptococcus pyogenes mgas10270
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: LIGASE Ligands: ZN, CIT, TRS, PEG |
 |
Crystal Structure Of The Murt/Gatd Enzyme Complex From Streptococcus Pyogenes With Bound Amppnp
Organism: Streptococcus pyogenes mgas10270
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: LIGASE Ligands: ZN, MG, ANP, GOL |
 |
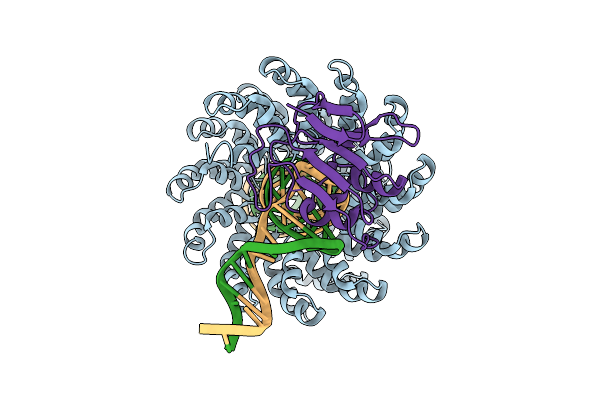
Organism: Xanthomonas, Burkholderia cenocepacia h111, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Organism: Xanthomonas, Burkholderia cenocepacia h111, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Organism: Cohnella sp. ov330
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: PROTEIN FIBRIL |
 |
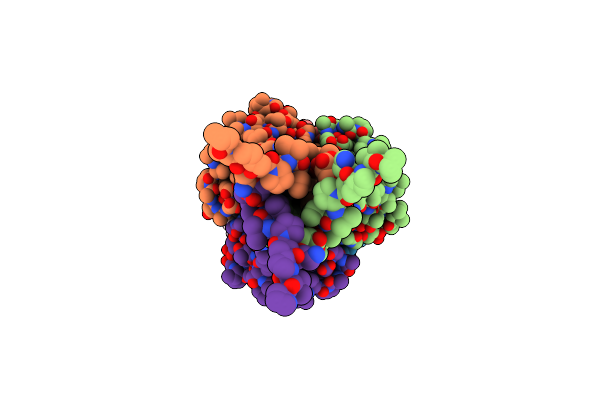
Organism: Paracandidimonas lactea
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: RNA Ligands: MG, K |
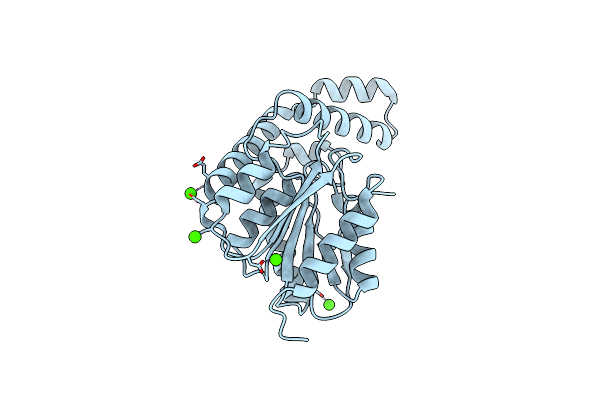 |
Fphh, Staphylococcus Aureus Fluorophosphonate-Binding Serine Hydrolases H, Apo Crystal Form 2
Organism: Staphylococcus aureus usa300-ca-263
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2025-01-08 Classification: HYDROLASE Ligands: CA |
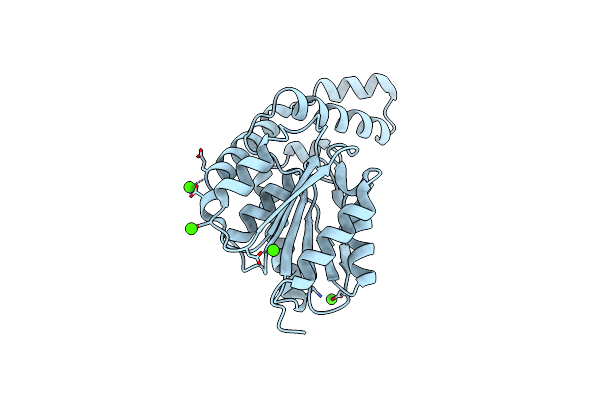 |
Fphh, Staphylococcus Aureus Fluorophosphonate-Binding Serine Hydrolases H, Apo Form 2 At Room Temperature
Organism: Staphylococcus aureus usa300-ca-263
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-01-08 Classification: HYDROLASE Ligands: CA |
 |
Fphi, Staphylococcus Aureus Fluorophosphonate-Binding Serine Hydrolases I, Apo Form At Room Temperature
Organism: Staphylococcus aureus usa300-ca-263
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-01-08 Classification: HYDROLASE Ligands: CA |
 |
Fphe, Staphylococcus Aureus Fluorophosphonate-Binding Serine Hydrolases E, Borolane-Based Compound Q41 Bound
Organism: Staphylococcus aureus subsp. aureus usa300
Method: X-RAY DIFFRACTION Release Date: 2024-11-20 Classification: HYDROLASE Ligands: XPU, CA |
 |
Organism: Achromobacter denitrificans nbrc 15125
Method: X-RAY DIFFRACTION Release Date: 2024-11-06 Classification: TRANSCRIPTION Ligands: ZN |
 |
Organism: Achromobacter denitrificans nbrc 15125
Method: X-RAY DIFFRACTION Release Date: 2024-11-06 Classification: TRANSCRIPTION Ligands: CL, 2HG, ZN |
 |
Fphe, Staphylococcus Aureus Fluorophosphonate-Binding Serine Hydrolases E, Boronic Acid-Based Compound Y43 Bound
Organism: Staphylococcus aureus subsp. aureus usa300
Method: X-RAY DIFFRACTION Release Date: 2024-10-23 Classification: HYDROLASE Ligands: WS8 |
 |
Fphe, Staphylococcus Aureus Fluorophosphonate-Binding Serine Hydrolases E, Boronic Acid-Based Compound Z27 Bound
Organism: Staphylococcus aureus subsp. aureus usa300
Method: X-RAY DIFFRACTION Release Date: 2024-10-16 Classification: HYDROLASE Ligands: WKK, CA |
 |
Organism: Escherichia coli k-12, Guillardia theta
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: ZN |
 |
Organism: Escherichia coli k-12, Guillardia theta, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: ZN |
 |
Organism: Escherichia coli k-12, Guillardia theta, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: ZN |
 |
Organism: Guillardia theta
Method: ELECTRON MICROSCOPY Resolution:2.71 Å Release Date: 2024-09-04 Classification: MEMBRANE PROTEIN Ligands: RET, PC1 |

