Search Count: 169
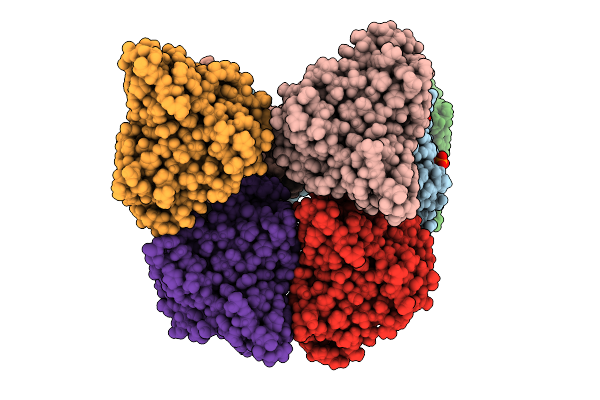 |
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-12-24 Classification: OXIDOREDUCTASE Ligands: CU, PO4 |
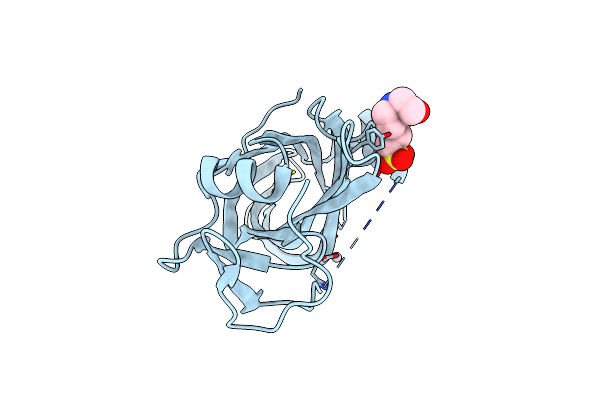 |
Crystal Structure Of Protein D: Defoliating Toxin Form Fusarium Oxysporum F.Sp. Vasinfectum
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2025-05-28 Classification: TOXIN Ligands: EPE, HG |
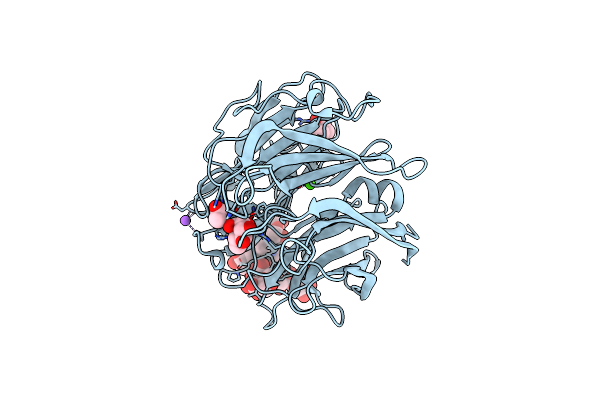 |
X-Ray Structure Of A L-Rhamnose-Alpha-1,4-D-Glucuronate Lyase From Fusarium Oxysporum 12S, L-Rha Complex At 100K
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2024-01-24 Classification: LYASE Ligands: RAM, ACT, TRS, CA, NA |
 |
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2023-11-29 Classification: HYDROLASE Ligands: RBH, XYS, YLL |
 |
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2023-11-29 Classification: HYDROLASE |
 |
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2023-11-22 Classification: HYDROLASE |
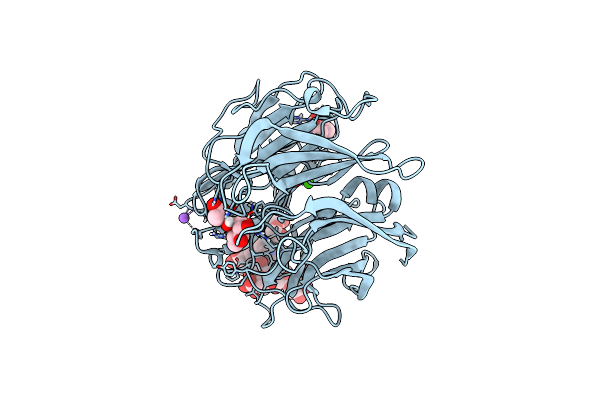 |
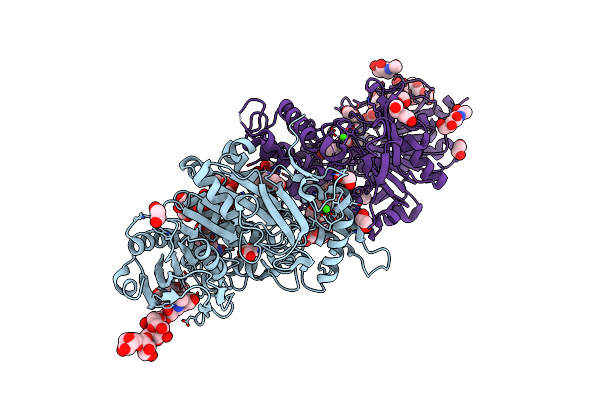
Neutron Structure Of A L-Rhamnose-Alpha-1,4-D-Glucuronate Lyase From Fusarium Oxysporum 12S, L-Rha Complex
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.25 Å, 1.8 Å Release Date: 2023-08-09 Classification: LYASE Ligands: RAM, ACT, TRS, CA, NA |
 |
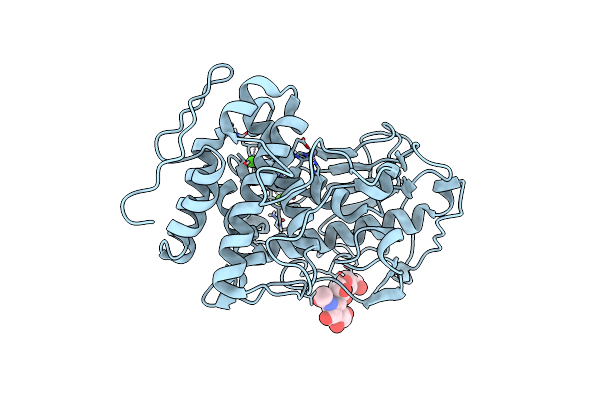
The Crystal Structure Of A Feruloyl Esterase C From Fusarium Oxysporum In Complex With P-Coumaric Acid
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2023-07-05 Classification: HYDROLASE Ligands: HC4, NAG, EDO, CA, PG4, P6G, PEG |
 |
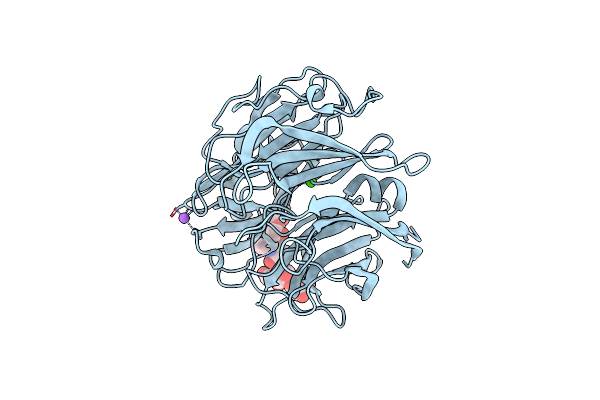
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2022-12-21 Classification: RECOMBINATION Ligands: ZN, CA |
 |
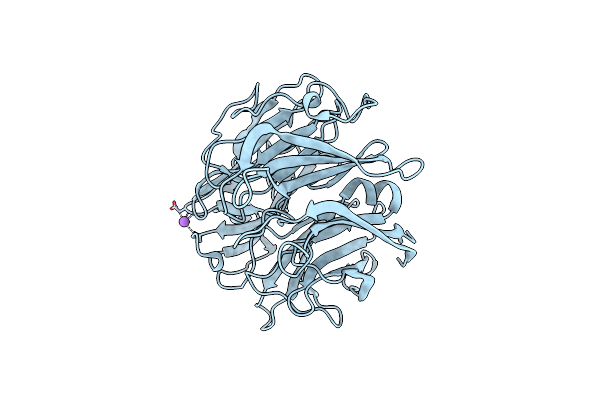
Crystal Structure Of A L-Rhamnose-Alpha-1,4-D-Glucuronate Lyase From Fusarium Oxysporum 12S, Ligand Free Form
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-08-04 Classification: LYASE Ligands: CA, NA |
 |
Crystal Structure Of A L-Rhamnose-Alpha-1,4-D-Glucuronate Lyase From Fusarium Oxysporum 12S, N247A N-Glycan Free Form
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-08-04 Classification: LYASE |
 |
Crystal Structure Of A L-Rhamnose-Alpha-1,4-D-Glucuronate Lyase From Fusarium Oxysporum 12S, L-Rha Complex
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-08-04 Classification: LYASE Ligands: RAM, ACT, NA |
 |
Crystal Structure Of A L-Rhamnose-Alpha-1,4-D-Glucuronate Lyase From Fusarium Oxysporum 12S, H105F Rha-Glca Complex
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2021-08-04 Classification: LYASE Ligands: TRS, NAG, SO4 |
 |
Structure Of Reaction Intermediate Of Cytochrome P450 No Reductase (P450Nor) Determined By Xfel
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-05-19 Classification: OXIDOREDUCTASE Ligands: HEM, NO, GOL |
 |
Crystal Structure Of A Novel 4-O-Alpha-L-Rhamnosyl-Beta-D-Glucuronidase From Fusarium Oxysporum 12S, Ligand-Free Form
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2021-03-17 Classification: HYDROLASE Ligands: NAG |
 |
Crystal Structure Of A Novel 4-O-Alpha-L-Rhamnosyl-Beta-D-Glucuronidase From Fusarium Oxysporum 12S - Rha-Glca Complex
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2021-03-17 Classification: HYDROLASE Ligands: NAG, MAN |
 |
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2020-08-19 Classification: HYDROLASE |
 |
Crystal Structure Of 2, 3-Dihydroxybenzoic Acid Decarboxylase From Fusarium Oxysporum
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-07-15 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN, GOL |
 |
Crystal Structure Of 2, 3-Dihydroxybenzoic Acid Decarboxylase From Fusarium Oxysporum In Complex With Catechol
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2020-07-15 Classification: BIOSYNTHETIC PROTEIN Ligands: CAQ, ZN |
 |
Crystal Structure Of 2, 3-Dihydroxybenzoic Acid Decarboxylase From Fusarium Oxysporum In Complex With 2,5-Dhba
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-07-15 Classification: BIOSYNTHETIC PROTEIN Ligands: GTQ, ZN |

