Search Count: 2,977
 |
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: OXIDOREDUCTASE Ligands: CU, PO4 |
 |
Cryo-Em Structure Of Gp77 Within The In Vitro Reconstituted Razr:Gp77 Complex
Organism: Escherichia phage secphi27
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN |
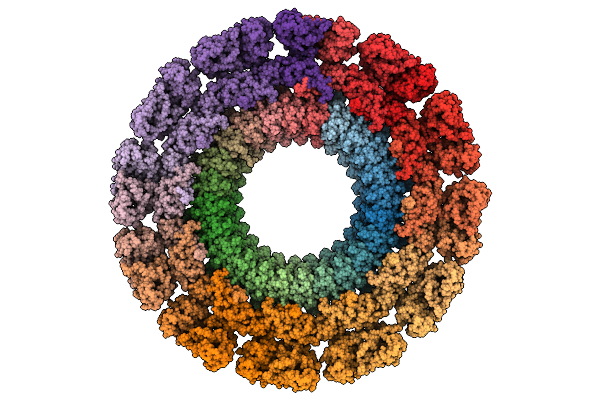 |
Cryo-Em Map Of The In Vitro Reconstituted Razr:Gp77 Complex With Alphafold-Predicted Models Fitted Into The Density.
Organism: Escherichia phage secphi27, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN Ligands: ZN |
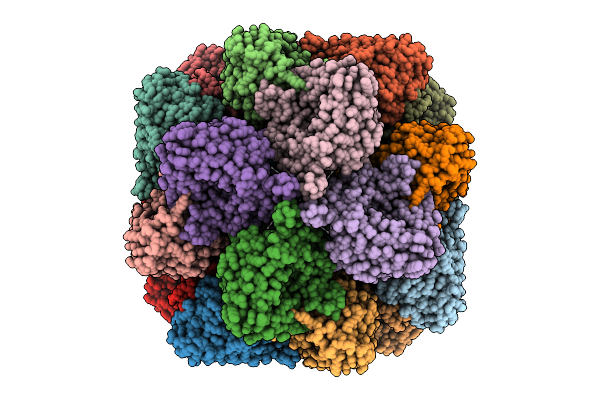 |
Cryo-Em Structure Of The Multimeric Phosphoenolpyruvate Binding Domain Of Staphylothermus Marinus Phosphoenolpyruvate Synthase
Organism: Staphylothermus marinus f1
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: CYTOSOLIC PROTEIN Ligands: MG |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: ANTIMICROBIAL PROTEIN Ligands: CL, GOL, EDO, PEG |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: ANTIMICROBIAL PROTEIN Ligands: NXL, A1IY4, CL, EDO, 2PE |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: ANTIMICROBIAL PROTEIN Ligands: OP0, EDO, GOL, PEG, CL, DMS |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: ANTIMICROBIAL PROTEIN Ligands: OP0, EDO, GOL, A1IYS, CL, 2PE |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE |
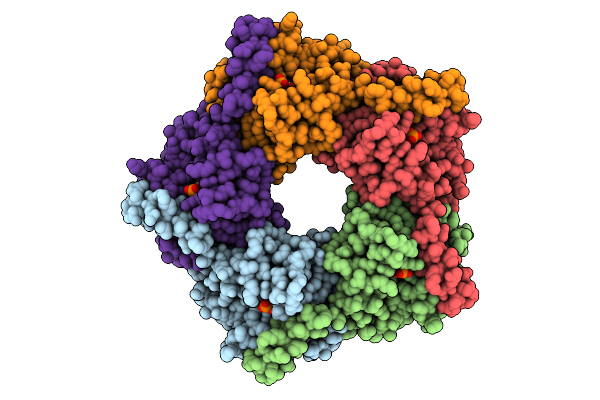 |
X-Ray Structure Of Enterobacter Cloaca Transaldolase In Complex With D-Fructose-6-Phosphate.
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: F6R |
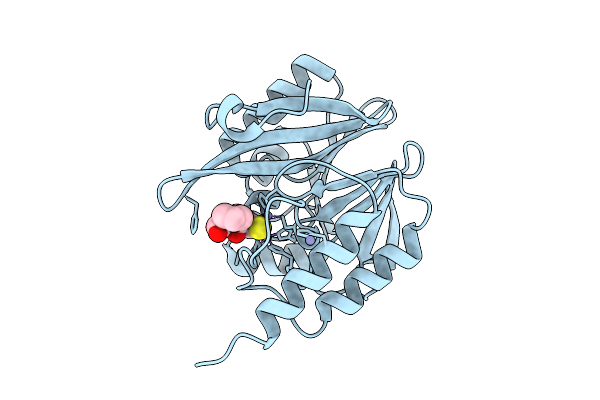 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE Ligands: ZN, MCO |
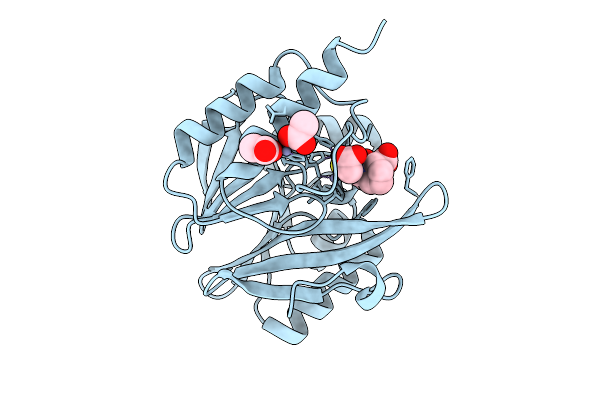 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE Ligands: ZN, X8Z, ACT |
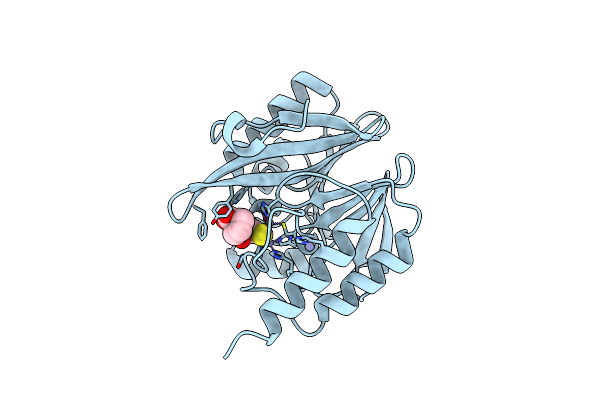 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE Ligands: ZN, MCO |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE Ligands: ZN, X8Z, MG |
 |
Crystal Structure Of Enterobacter Cloacae Ycdy, A Member Of The Redox Enzyme Maturation Protein Family
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: CHAPERONE Ligands: CA |
 |
Crystal Structure Of Protein D: Defoliating Toxin Form Fusarium Oxysporum F.Sp. Vasinfectum
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2025-05-28 Classification: TOXIN Ligands: EPE, HG |
 |
Organism: Burkholderia pyrrocinia, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2025-04-23 Classification: HYDROLASE |
 |
Organism: Brevibacillus
Method: X-RAY DIFFRACTION Release Date: 2025-01-01 Classification: BIOSYNTHETIC PROTEIN Ligands: ADP, A1LX0, MG, TPP |
 |
Organism: Brevibacillus
Method: X-RAY DIFFRACTION Release Date: 2024-11-20 Classification: BIOSYNTHETIC PROTEIN Ligands: TPP, ADP, ENO, MG |
 |
Organism: Brevibacillus
Method: X-RAY DIFFRACTION Release Date: 2024-11-20 Classification: BIOSYNTHETIC PROTEIN Ligands: ADP, TPP, MG |

