Search Count: 83
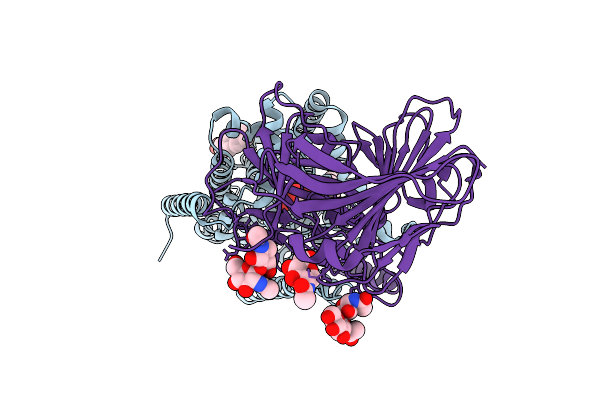 |
Cryo-Em Structure Of Candida Glabrata Gpi Mannosyltransferase I Bound To Dol-P-Man
Organism: Nakaseomyces glabratus
Method: ELECTRON MICROSCOPY Resolution:3.48 Å Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: MJC, Y01, NAG |
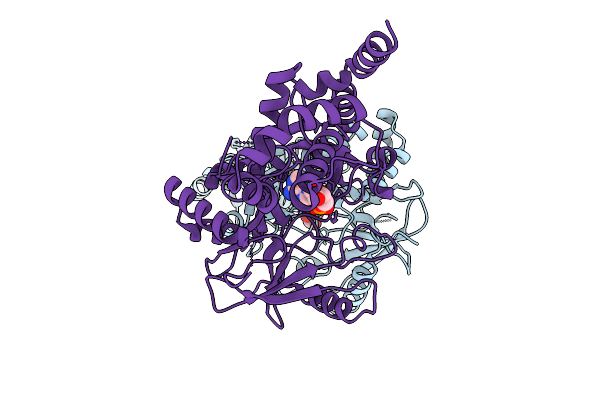 |
The Crystal Structure Of The Adenyly Transferase Domain Of Candida Glabrata Trna Ligase
Organism: Nakaseomyces glabratus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2025-09-17 Classification: LIGASE Ligands: AMP, CL |
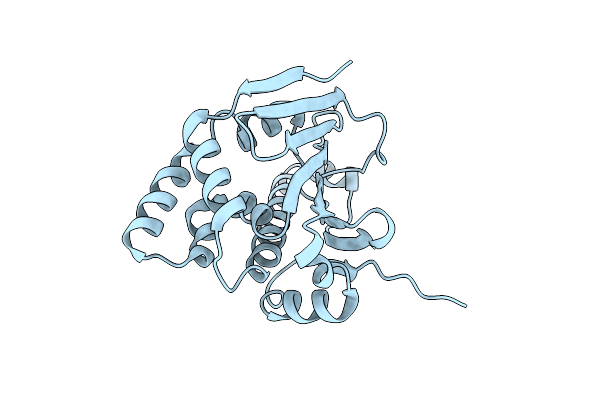 |
Crystal Structure Of The Nucleotide-Binding Domain Of Candida Glabrata Hsp90
Organism: Candida glabrata
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-03-05 Classification: CHAPERONE |
 |
Crystal Structure Of Candida Glabrata Bdf1 Bromodomain 1 In The Unbound State
Organism: Nakaseomyces glabratus
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2024-10-30 Classification: TRANSCRIPTION |
 |
Crystal Structure Of Candida Glabrata Bdf1 Bromodomain 1 Bound To A Pyrazole Ligand
Organism: Nakaseomyces glabratus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2024-10-30 Classification: TRANSCRIPTION Ligands: Y7F, SO4 |
 |
Crystal Structure Of Candida Glabrata Bdf1 Bromodomain 1 Bound To A Phenyltriazine Ligand
Organism: Nakaseomyces glabratus
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2024-10-30 Classification: TRANSCRIPTION Ligands: Y78, PO4, K |
 |
Crystal Structure Of Candida Glabrata Bdf1 Bromodomain 2 In The Unbound State
Organism: Nakaseomyces glabratus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2024-10-30 Classification: TRANSCRIPTION Ligands: MPD, GOL, ACT |
 |
Organism: Nakaseomyces glabratus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-10-30 Classification: TRANSCRIPTION Ligands: GOL, SO4, 1GH |
 |
Crystal Structure Of Candida Glabrata Bdf1 Bromodomain 2 Bound To A Pyridoindole Ligand
Organism: Nakaseomyces glabratus
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-10-30 Classification: TRANSCRIPTION Ligands: Y7B |
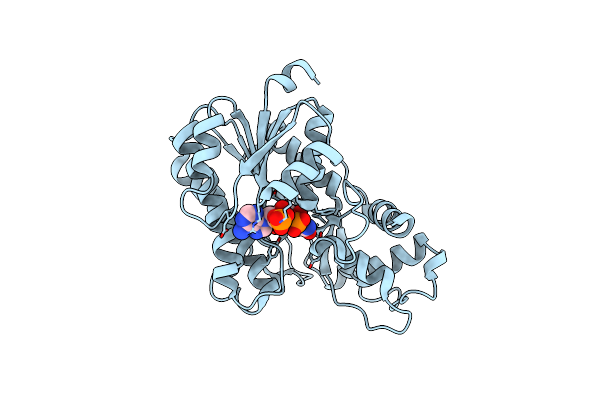 |
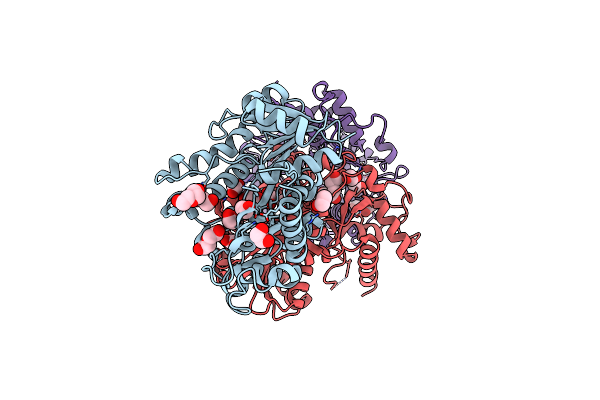
Structure Of Alcohol Dehydrogenase From Candida Glabrata(Cgadh)Complexed With Nadph
Organism: [candida] glabrata
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-08-02 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
Organism: [candida] glabrata
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-08-02 Classification: OXIDOREDUCTASE Ligands: EDO, PEG, CIT |
 |
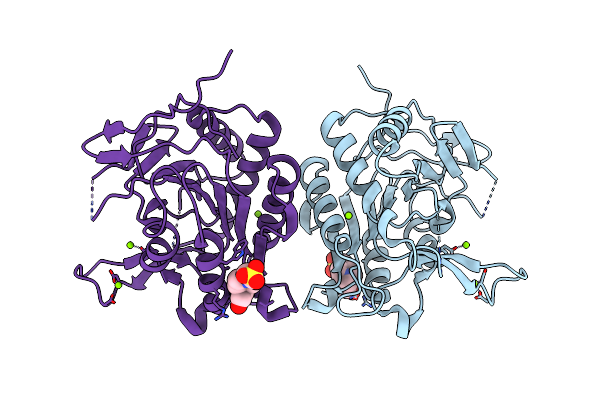
Organism: [candida] glabrata
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2023-01-18 Classification: CELL ADHESION |
 |
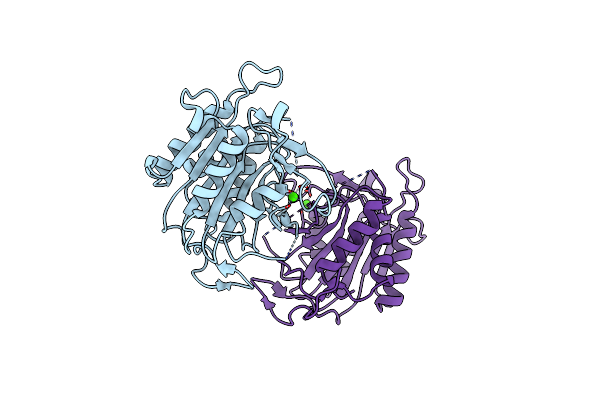
Organism: [candida] glabrata
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-01-11 Classification: APOPTOSIS Ligands: EPE, MG |
 |
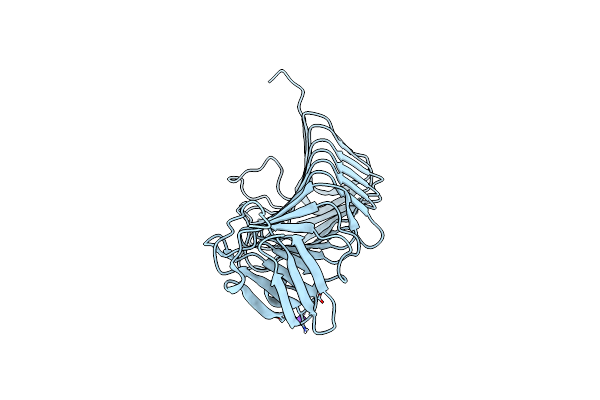
Organism: [candida] glabrata
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-01-11 Classification: APOPTOSIS Ligands: CA, CL |
 |
Crystal Structure Of The Awp3B (Adhesin-Like Wall Protein 3B) A-Domain From Candida Glabrata
Organism: Candida glabrata
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2021-12-15 Classification: CELL ADHESION Ligands: CL, NA |
 |
Crystal Structure Of The Awp3B (Adhesin-Like Wall Protein 3B) A-Domain From Candida Glabrata Showing A Gadolinium Cluster
Organism: Candida glabrata
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2021-12-15 Classification: CELL ADHESION Ligands: GD |
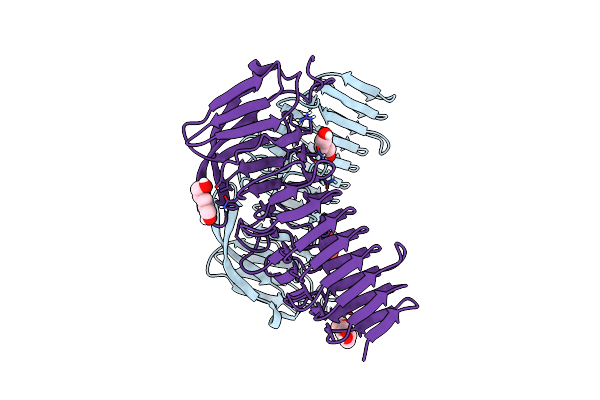 |
Crystal Structure Of The Awp1 (Adhesin-Like Wall Protein 1) A-Domain From Candida Glabrata
Organism: [candida] glabrata
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-12-15 Classification: CELL ADHESION Ligands: 9JE |
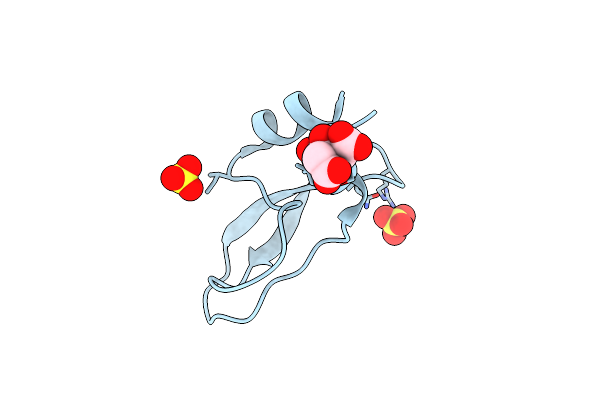 |
A Lid Blocking Mechanism Of A Cone Snail Toxin Revealed At The Atomic Level
Organism: Conus cocceus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-04-14 Classification: TOXIN Ligands: SO4, CIT |
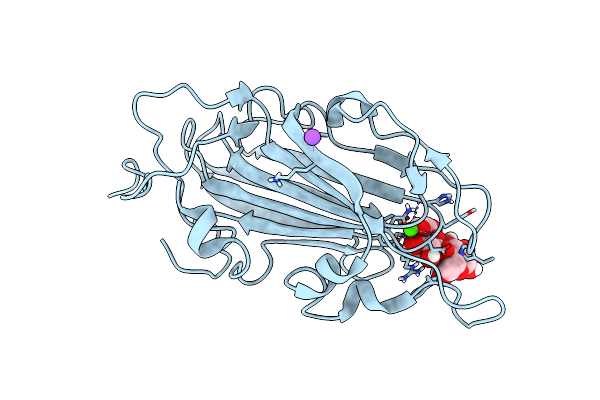 |
Crystal Structure Of Subtype-Switched Epithelial Adhesin 1 To 9 A Domain (Epa1-Cbl2Epa9) From Candida Glabrata In Complex With Beta-Lactose
Organism: Candida glabrata
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2020-07-22 Classification: SUGAR BINDING PROTEIN Ligands: CA, CL, NA |
 |
Tsh2 Domain Of Transcription Elongation Factor Spt6 Complexed With Tyrosine Phosphorylated Ctd
Organism: Candida glabrata
Method: SOLUTION NMR Release Date: 2020-07-15 Classification: GENE REGULATION |

