Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Gelsemium sempervirens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: PLANT PROTEIN Ligands: PEG, SO4, EDO |
 |
Organism: Actinomadura luzonensis
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, EDO |
 |
N-Hydroxylamine Dehydratase (Nohd) T98A/K167A Mutant Crystal Structure With Heme And N-Hydroxylated Ornithine (2.5H Soak)
Organism: Actinomadura luzonensis
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, HEM, ONH |
 |
N-Hydroxylamine Dehydratase (Nohd) T98A/K167A Mutant Crystal Structure With Heme And N-Hydroxylated Ornithine (5H Soak)
Organism: Actinomadura luzonensis
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, ONH |
 |
N-Hydroxylamine Dehydratase (Nohd) R144Y/V96A (A1/A2) Mutant Crystal Structure With Heme
Organism: Actinomadura luzonensis
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, GOL, SO4 |
 |
N-Hydroxylamine Dehydratase (Nohd) H2F/F4P/P5S/R6Y/A59N/R144A (P1/P2/A1) Mutant Crystal Structure With Heme
Organism: Actinomadura luzonensis
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, EDO |
 |
N-Hydroxylamine Dehydratase (Nohd) H2F/F4P/P5S/R6Y/R144Y/V96A (P1/A1/A2) Mutant Crystal Structure With Heme
Organism: Actinomadura luzonensis
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, GOL, SO4 |
 |
N-Hydroxylamine Dehydratase (Nohd) A59N/R144Y/V96A (P2/A1/A2) Mutant Crystal Structure With Heme
Organism: Actinomadura luzonensis
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM |
 |
N-Hydroxylamine Dehydratase (Nohd) H2F/F4P/P5S/R6Y/A59N/R144Y/V96A (P1/P2/A1/A2) Mutant Crystal Structure With Heme
Organism: Actinomadura luzonensis
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, EDO |
 |
N-Hydroxylamine Dehydratase (Nohd) H2F/F4P/P5S/R6Y/A59N/R144Y/V96A/D172E/L172Q/D173P/R176V (P1/P2/A1/A2/A3) Mutant Crystal Structure With Heme And N-Hydroxylated Ornithine
Organism: Actinomadura luzonensis
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, BTB, ONH |
 |
Organism: Campylobacter jejuni subsp. jejuni serotype o:23/36 (strain 81-176)
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: STRUCTURAL PROTEIN |
 |
Organism: Bacterium hr29
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: HYDROLASE |
 |
Organism: Sarbecovirus sp. rhgb07
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Pyrodictium abyssi
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: CELL ADHESION |
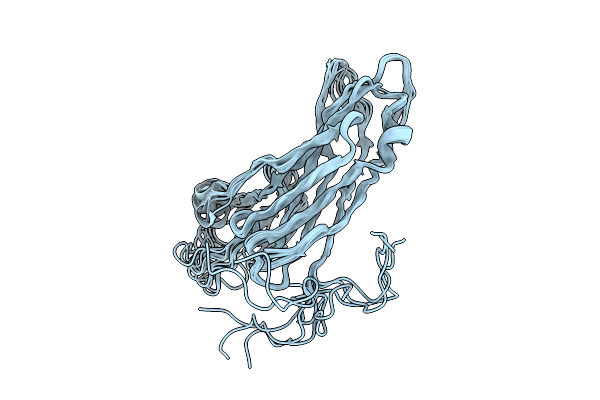 |
Organism: Pyrodictium abyssi
Method: SOLUTION NMR Release Date: 2025-08-20 Classification: CELL ADHESION |
 |
Organism: Cochliomyia hominivorax
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE |
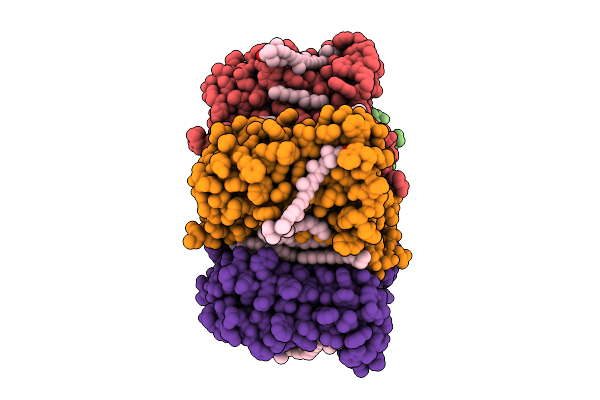 |
Cryo-Em Structure Of The Zeaxanthin-Bound Light-Driven Proton Pumping Rhodopsin, Nm-R1
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, R16, D12, K3I |
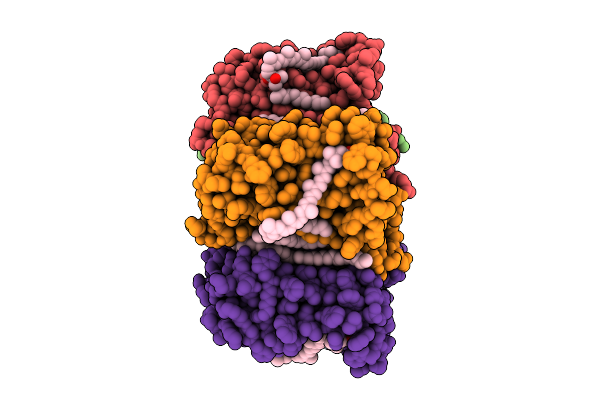 |
Cryo-Em Structure Of The Myxol-Bound Light-Driven Proton Pumping Rhodopsin, Nm-R1
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, R16, D12, A1L4O |
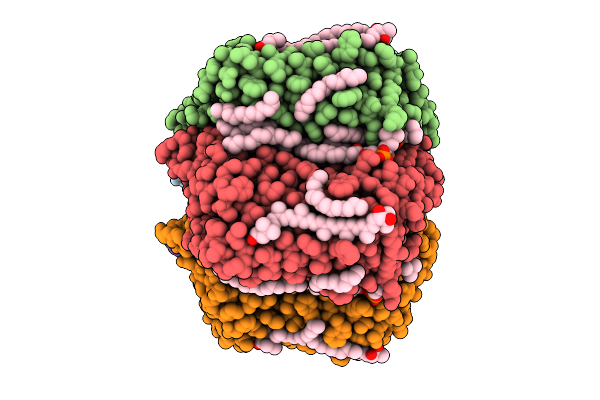 |
Cryo-Em Structure Of The Myxol-Bound Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, A1L4O, CL, PC1, PLC, R16, 8K6, D12, C14, D10 |
 |
Cryo-Em Structure Of The Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, CL, PC1, PLC, D12, R16, 8K6, C14 |