Search Count: 3,197
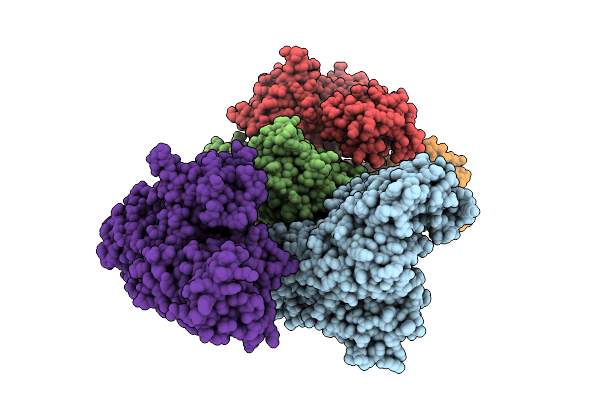 |
Crystal Structure Of A Wild-Type Tagose Isomerase (Tst4Ease Wt) From Thermotogota Bacterium
Organism: Thermotogota bacterium
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2025-12-24 Classification: ISOMERASE |
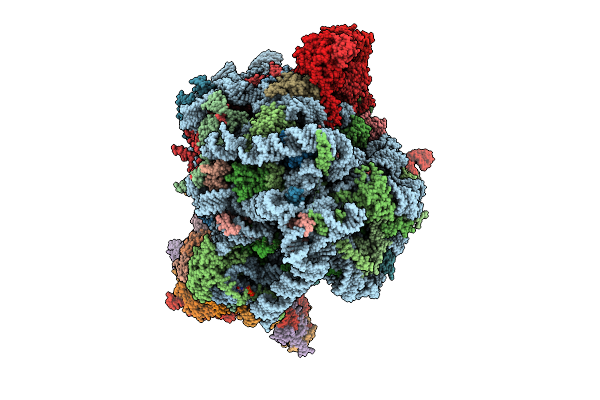 |
Human Quaternary Complex Of A Translating 80S Ribosome, Nac, Metap1 And Natd
Organism: Homo sapiens, Saccharomyces cerevisiae s288c, Aequorea victoria, Brachypodium distachyon
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: ZN, COA, GTP |
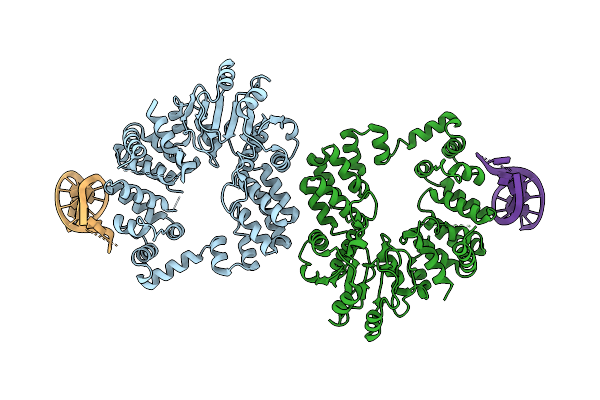 |
Organism: Thermus phage philo
Method: ELECTRON MICROSCOPY Resolution:3.81 Å Release Date: 2025-12-10 Classification: RNA BINDING PROTEIN/RNA |
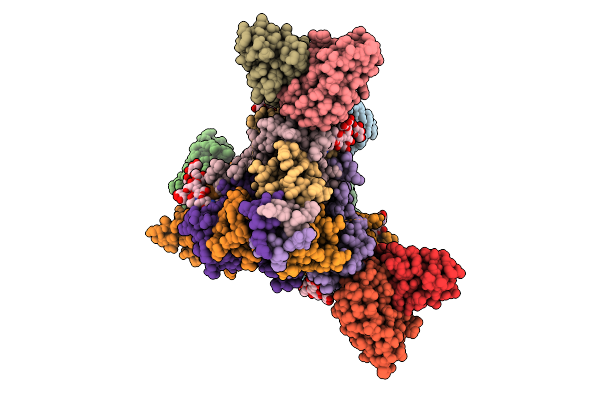 |
A Broadly-Neutralizing Antibody Against Ebolavirus Glycoprotein That Can Potentiate The Breadth And Neutralization Potency Of Other Anti-Glycoprotein Antibodies
Organism: Oryctolagus cuniculus, Ebolavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Thermus phage philo
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2025-10-01 Classification: RNA BINDING PROTEIN Ligands: CA |
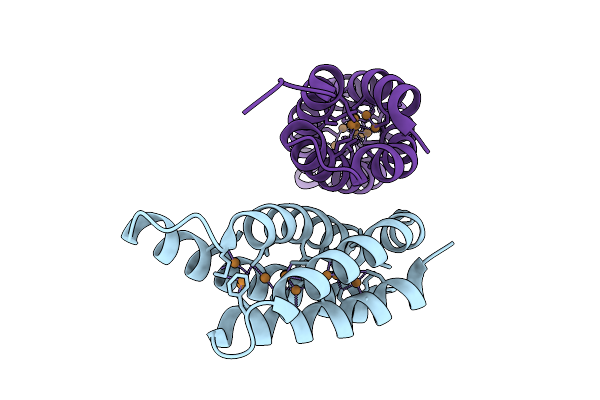 |
Organism: Methylosinus trichosporium ob3b
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-09-24 Classification: METAL BINDING PROTEIN Ligands: CU1 |
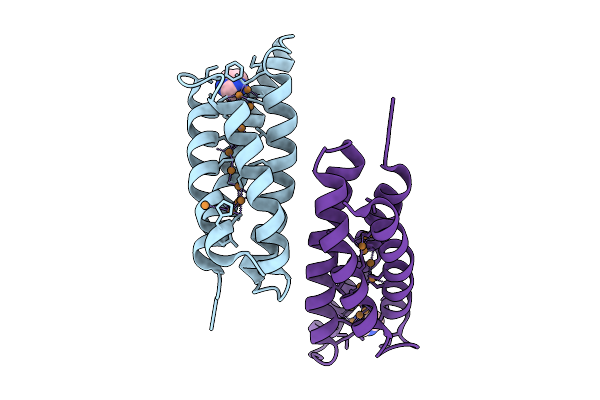 |
Organism: Methylosinus trichosporium ob3b
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2025-09-24 Classification: METAL BINDING PROTEIN Ligands: CU1, CU, IMD |
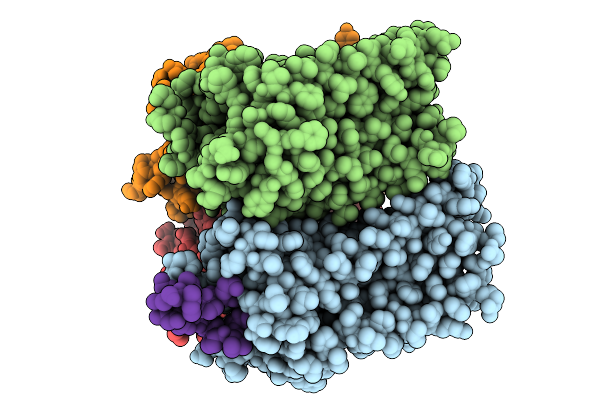 |
Organism: Homo sapiens, Virus-associated rnas
Method: ELECTRON MICROSCOPY Resolution:3.57 Å Release Date: 2025-09-10 Classification: SIGNALING PROTEIN |
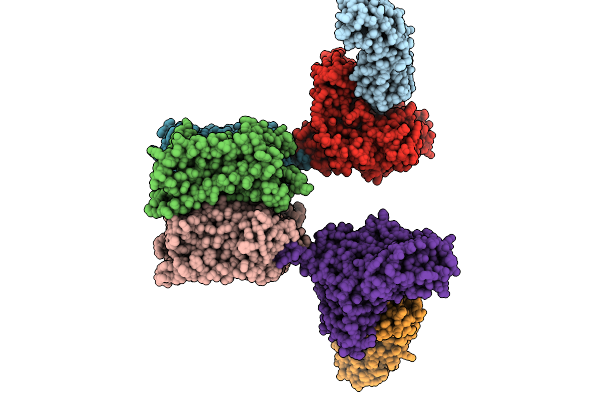 |
Organism: Homo sapiens, Virus-associated rnas
Method: ELECTRON MICROSCOPY Resolution:5.65 Å Release Date: 2025-09-10 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of An Agose Isomerase Mutant 5 (Tst4Ease M5) From Thermotogota Bacterium With Zn
Organism: Thermotogota bacterium
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2025-07-09 Classification: ISOMERASE Ligands: ZN |
 |
Crystal Structure Of A Wild-Type Tagose Isomerase (Tst4Ease Wt) From Thermotogota Bacterium Complex With Zn
Organism: Thermotogota bacterium
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2025-07-09 Classification: ISOMERASE Ligands: ZN |
 |
Chimeric Adenosine Deaminase Growth Factor (Adgf) In Complex With Pentostatin
Organism: Aplysia californica, Helix pomatia
Method: X-RAY DIFFRACTION Resolution:3.28 Å Release Date: 2025-06-25 Classification: HYDROLASE Ligands: NAG, EDO, ZN, DCF |
 |
Organism: Helix pomatia
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-06-18 Classification: HYDROLASE Ligands: EDO, NAG, BTB, GOL, PEG, ZN, PGE |
 |
Helix Pomatia Amp Deaminase (Hpampd) With Unknown Density In The Active Site
Organism: Helix pomatia
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2025-06-18 Classification: HYDROLASE Ligands: NAG, ZN, EDO, BTB, GOL, PO4, ACT |
 |
Organism: Helix pomatia
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2025-06-18 Classification: HYDROLASE Ligands: NAG, ZN, EDO, BTB, DCF |
 |
Helix Pomatia Amp Deaminase (Hpampd) In Complex With Pentostatin Monophosphate (Pmp)
Organism: Helix pomatia
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2025-06-18 Classification: HYDROLASE Ligands: EDO, NAG, BTB, GOL, ZN, A1CDW, PEG |
 |
Cu-Bound Tetrameric Copper Storage Protein 1 With Anti-Rhodopsin 1D4 Epitope
Organism: Methylosinus trichosporium ob3b
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: METAL BINDING PROTEIN |
 |
Ligand Binding Domain Of Roseburia Intestinalis L1-82 Uracil Chemoreceptor (Dcache) In Complex With Uracil And Acetate
Organism: Roseburia intestinalis l1-82
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2025-03-12 Classification: SIGNALING PROTEIN Ligands: URA, ACT, EDO |
 |
Cryoem Structure Of Bont-Ntnh-Orfx2 Complex From Clostridium Botulinum E1, Major Class
Organism: Clostridium botulinum e1 str. 'bont e beluga'
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: TOXIN |
 |
Cryoem Structure Of Bont-Ntnh-Orfx2 Complex From Clostridium Botulinum E1, Minor Class
Organism: Clostridium botulinum e1 str. 'bont e beluga'
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: TOXIN |

