Search Count: 14
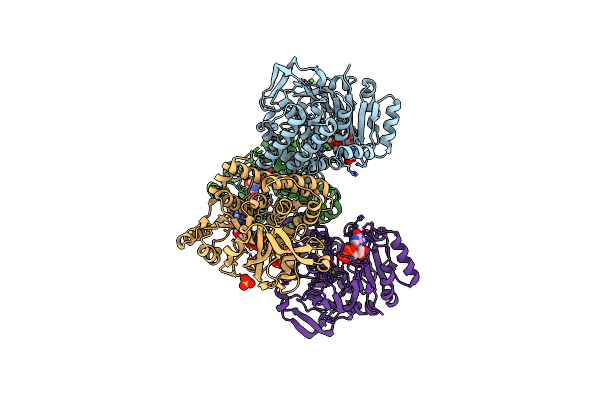 |
Structural Insight Into A Metal-Dependent Mutase Mtdl Revealing An Arginine Residue Covalently Mediated Interconversion Between Nucleotide-Based Furanose And Pyranose
Organism: Marinactinospora thermotolerans
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-07-26 Classification: BIOSYNTHETIC PROTEIN Ligands: GDP, MG, SO4 |
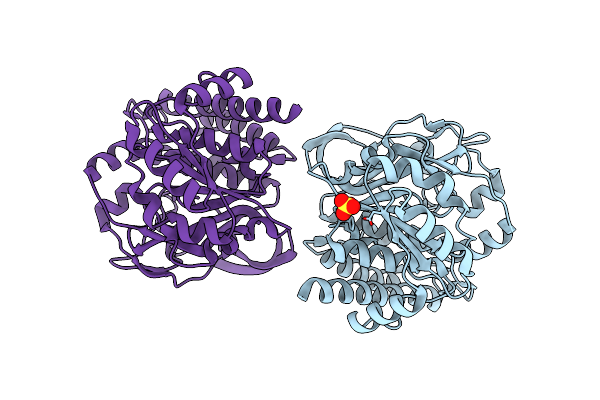 |
Structural Insight Into A Metal-Dependent Mutase Mtdl Revealing An Arginine Residue Covalently Mediated Interconversion Between Nucleotide-Based Furanose And Pyranose
Organism: Marinactinospora thermotolerans
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2023-07-26 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4 |
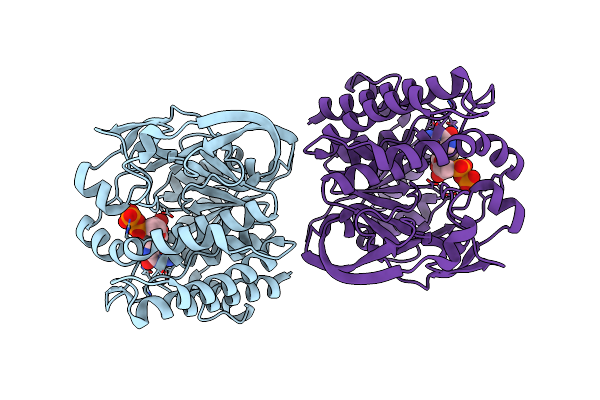 |
Organism: Marinactinospora thermotolerans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-03-29 Classification: TRANSFERASE Ligands: GDP |
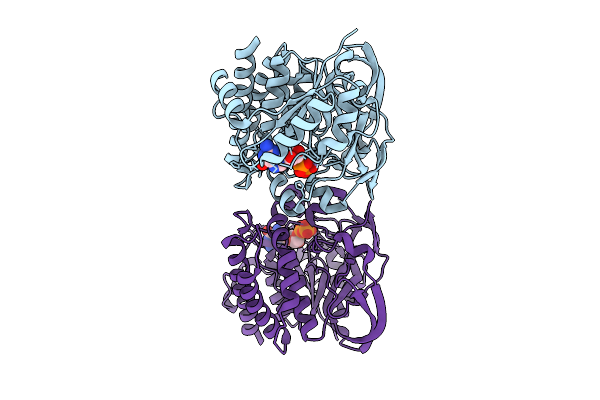 |
Organism: Marinactinospora thermotolerans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-03-29 Classification: TRANSFERASE Ligands: GDP, MN |
 |
Organism: Marinactinospora thermotolerans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-03-29 Classification: TRANSFERASE Ligands: GDG, MN |
 |
Organism: Marinactinospora thermotolerans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-03-29 Classification: TRANSFERASE Ligands: GFB, MN, GOL |
 |
Organism: Marinactinospora thermotolerans
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-03-29 Classification: TRANSFERASE Ligands: GRJ, MN |
 |
Organism: Marinactinospora thermotolerans
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-03-29 Classification: TRANSFERASE Ligands: MN |
 |
Crystal Structure Of A Cutinase Enzyme From Marinactinospora Thermotolerans Dsm45154 (606)
Organism: Marinactinospora thermotolerans
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2022-12-28 Classification: HYDROLASE |
 |
Structure Of Amide Bond Synthetase Mcba From Marinactinospora Thermotolerans
Organism: Marinactinospora thermotolerans
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2020-04-22 Classification: LIGASE Ligands: EQ2, AMP |
 |
Structure Of Amide Bond Synthetase Mcba K483A Mutant From Marinactinospora Thermotolerans
Organism: Marinactinospora thermotolerans
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-09-05 Classification: LIGASE Ligands: AMP, EQ2 |
 |
Crystal Structure Of The Indole Prenyltransferase Mpnd From Marinactinospora Thermotolerans
Organism: Marinactinospora thermotolerans
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-03-16 Classification: TRANSFERASE |
 |
Crystal Structure Of The Indole Prenyltransferase Mpnd Complexed With Indolactam V And Dmspp
Organism: Marinactinospora thermotolerans
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-03-16 Classification: TRANSFERASE Ligands: ILV, SO4, DST |
 |
Organism: Marinactinospora thermotolerans
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2015-10-28 Classification: LYASE Ligands: TRP |

