Search Count: 514
 |
X-Ray Structure Of The Zn-Dependent Receptor-Binding Domain Of Proteus Mirabilis Mr/P Fimbrial Adhesin Mrph
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2020-08-19 Classification: CELL ADHESION Ligands: ZN, TLA |
 |
X-Ray Structure Of The Zn-Dependent Receptor-Binding Domain Of Proteus Mirabilis Mr/P Fimbrial Adhesin Mrph
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-08-19 Classification: CELL ADHESION Ligands: ZN, GLU |
 |
Crystal Structure Of The Acyl-Carrier-Protein Udp-N-Acetylglucosamine O-Acyltransferase Lpxa From Proteus Mirabilis
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2020-01-29 Classification: TRANSFERASE Ligands: SO4, PO3 |
 |
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2019-06-26 Classification: LYASE Ligands: SO4 |
 |
Organism: Proteus mirabilis hi4320
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2018-11-07 Classification: CELL ADHESION Ligands: GOL, PO4 |
 |
Receptor-Binding Domain Of Proteus Mirabilis Uroepithelial Cell Adhesin Ucad21-211
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-11-07 Classification: CELL ADHESION Ligands: CO |
 |
Receptor-Binding Domain Of Proteus Mirabilis Uroepithelial Cell Adhesin Ucad21-217
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-11-07 Classification: CELL ADHESION Ligands: SO4 |
 |
Substrate-Bound Outward-Open State Of A Na+-Coupled Sialic Acid Symporter Reveals A Novel Na+-Site
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-04-04 Classification: MEMBRANE PROTEIN |
 |
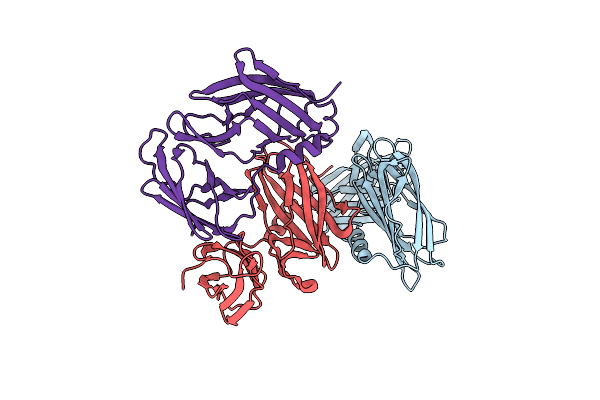
Substrate-Bound Outward-Open State Of A Na+-Coupled Sialic Acid Symporter Reveals A Novel Na+-Site
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2018-04-04 Classification: MEMBRANE PROTEIN |
 |
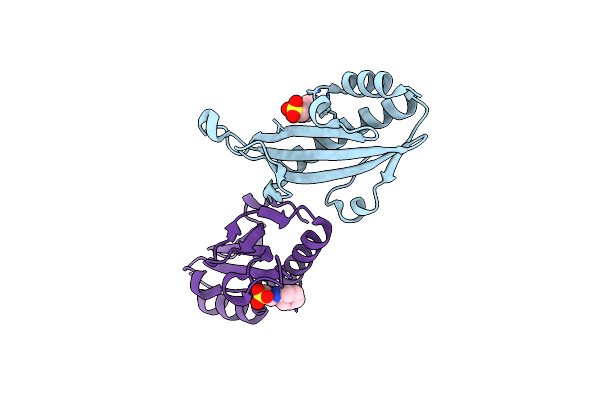
Crystal Structure Of The N-Terminal Periplasmic Domain Of Scsb From Proteus Mirabilis
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2018-03-07 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Universal Stress Protein E From Proteus Mirabilis In Complex With Udp-3-O-[(3R)-3-Hydroxytetradecanoyl]-N-Acetyl-Alpha-Glucosamine
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-11-26 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: U20, GOL, CL, SO4 |
 |
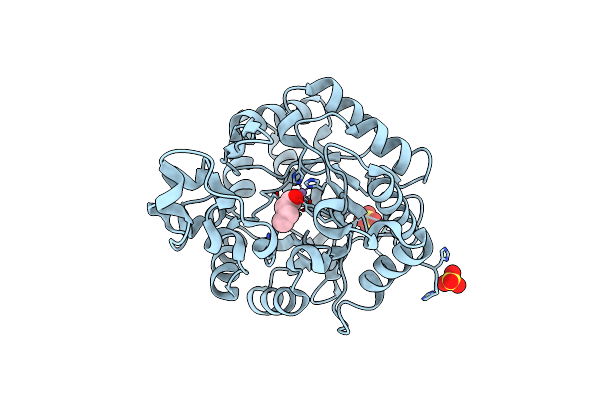
Crystal Structure Of Proteus Mirabilis Transcriptional Regulator Protein Crl At 1.95A Resolution
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2014-08-06 Classification: Transcription regulator Ligands: NHE |
 |
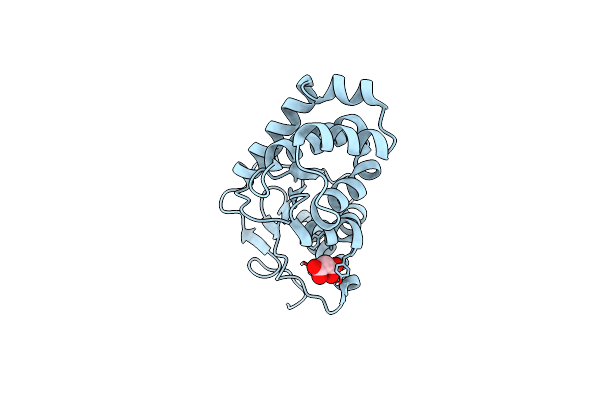
Crystal Structure Of Amidohydrolase Pmi1525 (Target Efi-500319) From Proteus Mirabilis Hi4320, A Complex With Butyric Acid And Manganese
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-07-30 Classification: HYDROLASE Ligands: MN, BUA, SO4 |
 |
Crystal Structure Of The Disulfide Oxidoreductase Dsba From Proteus Mirabilis
Organism: Proteus mirabilis hi4320
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2014-05-28 Classification: OXIDOREDUCTASE Ligands: MLI |
 |
Crystal Structure Of The Disulfide Oxidoreductase Dsba (S30Xxc33) Active Site Mutant From Proteus Mirabilis
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2014-05-28 Classification: OXIDOREDUCTASE Ligands: SCN |
 |
Complex Structure Of Proteus Mirablis Dsba (C30S) With A Non-Covalently Bound Peptide Pwatcds
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-05-28 Classification: OXIDOREDUCTASE/PEPTIDE Ligands: SCN |
 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-12-04 Classification: HYDROLASE |
 |
Structure Of A Curlin Genes Transcriptional Regulator Protein From Proteus Mirabilis Hi4320.
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-08-31 Classification: transcription regulator Ligands: SO4, EDO |
 |
Crystal Structure Of Amidohydrolase Pmi1525 (Target Efi-500319) From Proteus Mirabilis Hi4320
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2011-04-27 Classification: HYDROLASE Ligands: ZN, BEZ, CAC, SO4, UNL |
 |
The Crystal Structure Of A Universal Stress Protein E From Proteus Mirabilis Hi4320
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2010-09-22 Classification: structural genomics, unknown function Ligands: GOL, PEG, ACT, UNL |

