Search Count: 691
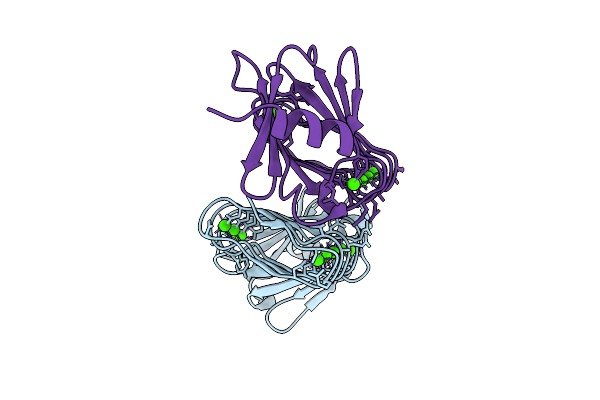 |
Rtx Domain Block V Of Adenylate Cyclase Toxin With Mutations D1533N, A1542N, D1560N, S1569N, D1587N, H1598N, H1608N
Organism: Bordetella pertussis
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TOXIN Ligands: CA |
 |
Cryo Em Structure Of Pertussis Toxin In Complex With Neutralizing Fab Pert-61
Organism: Bordetella pertussis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: TOXIN/Immune System |
 |
Cryo Em Structure Of Pertussis Toxin In Complex With Neutralizing Fab Pert-171
Organism: Bordetella pertussis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: TOXIN/Immune System |
 |
Cryo Em Structure Of Pertussis Toxin In Complex With Neutralizing Fab Pert-199
Organism: Bordetella pertussis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: TOXIN/Immune System |
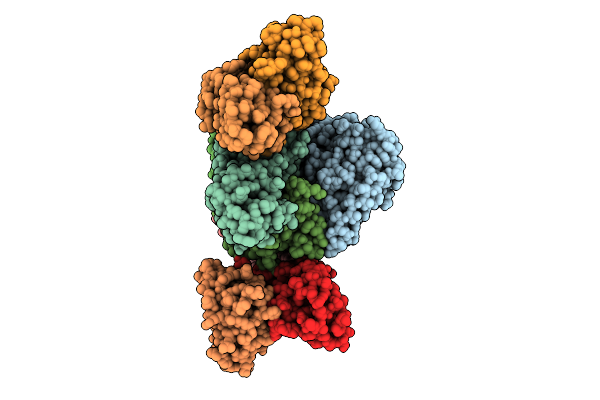 |
Cryo Em Structure Of Pertussis Toxin In Complex With Neutralizing Fab Pert-169 And Pert-203
Organism: Bordetella pertussis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: TOXIN/Immune System |
 |
Cryo-Em Structure Of Influenza B/Washington/02/2019 Virus Hemagglutinin In Complex With Single-Domain Antibody Hvhh-69.
Organism: Influenza b virus, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Influenza B/Washington/02/2019 Virus Neuraminidase In Complex With Single-Domain Antibody Hvhh-525.
Organism: Lama glama, Influenza b virus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
Organism: Influenza b virus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Parachlamydia sp., Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: TLA |
 |
Organism: Homo sapiens, Parachlamydia sp.
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: CIT |
 |
Genetiocally Detoxified Pertussis Toxin In Complex With Hu1B7 Fab And Hu11E6 Fab
Organism: Bordetella pertussis, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: TOXIN |
 |
Organism: Lactiplantibacillus paraplantarum
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: GC2, EDO, PEG |
 |
Organism: Clostridium beijerinckii
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: OXIDOREDUCTASE Ligands: ZN, SF4, 402 |
 |
Organism: Influenza b virus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Crystal Structure Of Apo-[Fefe]-Hydrogenase Cba5H From Clostridium Beijerinckii
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2024-12-18 Classification: OXIDOREDUCTASE Ligands: SF4, ZN, CL |
 |
Crystal Structure Of [Fefe]-Hydrogenase Cba5H From Clostridium Beijerinckii In Hinact State
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-12-18 Classification: OXIDOREDUCTASE Ligands: 402, SF4, ZN, CL |
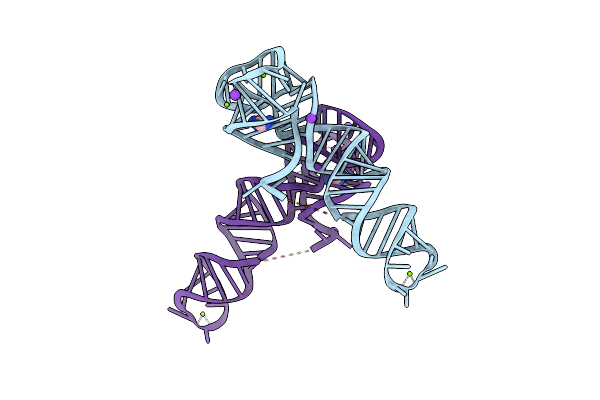 |
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-11-27 Classification: RNA Ligands: MG, 5AC, K |
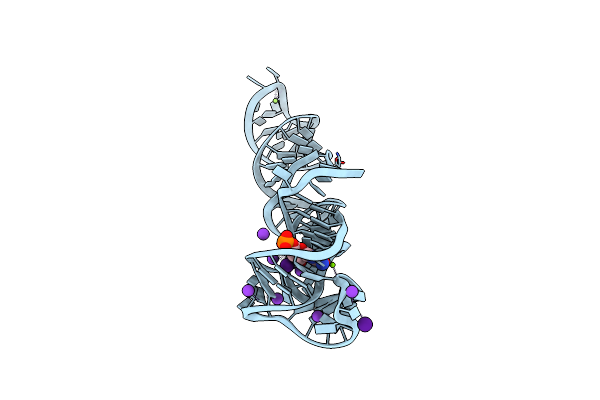 |
Cocrystal Structure Of Clostridium Beijerinckii Ztp Riboswitch With Zmp And Cs
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2024-11-27 Classification: RNA Ligands: CS, MG, AMZ, K |
 |
Organism: Myroides profundi
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ELECTRON TRANSPORT |
 |
Organism: Myroides profundi
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ELECTRON TRANSPORT Ligands: KEN |

