Search Count: 28,831
 |
Rtx Domain Block V Of Adenylate Cyclase Toxin With Mutations D1533N, A1542N, D1560N, S1569N, D1587N, H1598N, H1608N
Organism: Bordetella pertussis
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TOXIN Ligands: CA |
 |
Structure Of The Energy Converting Methyltransferase (Mtr) Of Methanosarcina Mazei In Complex With A Novel Protein Binder
Organism: Methanosarcina mazei go1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: A1JAP, L1P, A1JAO, NA, A1JAN |
 |
Structure Of The Energy Converting Methyltransferase (Mtr) Of Methanosarcina Mazei In Complex With A Novel Protein Binder
Organism: Methanosarcina mazei go1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: B13, A1JAP, L1P, A1JAO, NA, A1JAN |
 |
Crystal Structure Of A Glyceraldehyde-3-Phosphate Dehydrogenase From Neisseria Gonorrhoeae In Complex With Nad (P1 Form2)
Organism: Neisseria gonorrhoeae nccp11945
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: OXIDOREDUCTASE Ligands: NAD, NA |
 |
Crystal Structure Of Nucleoside-Diphosphate Kinase Cryptosporidium Parvum (Apo, Hexamer)
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSFERASE Ligands: EDO, CL |
 |
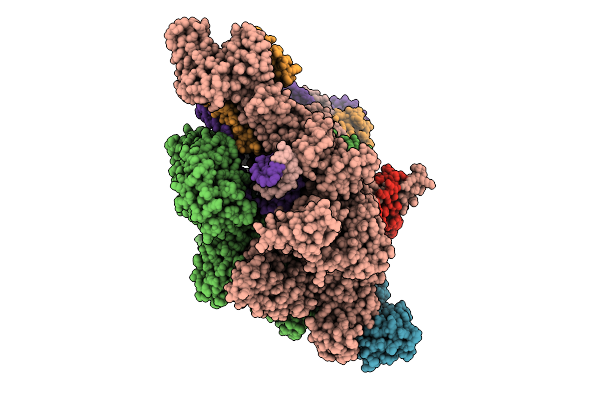
Organism: Thermus thermophilus hb8, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: MG, ZN, GTP, 2TM |
 |
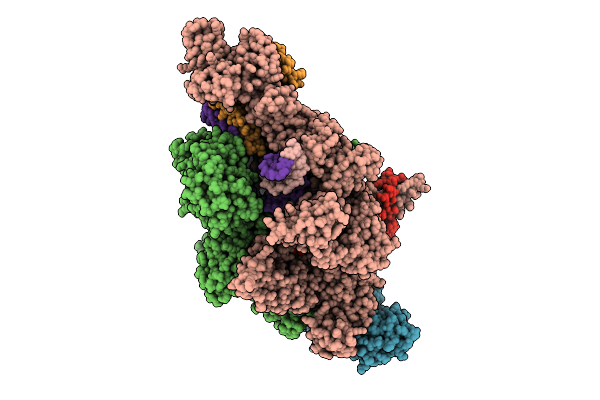
Organism: Thermus thermophilus hb8, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: MG, ZN, 2TM, G3A |
 |
Organism: Thermus thermophilus hb8, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: MG, ZN, 2TM, A1IF9 |
 |
Organism: Thermus thermophilus hb8, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: MG, B4P, ZN, G2P |
 |
Organism: Thermus thermophilus hb8, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: MG, ZN, 2TM, B4P |
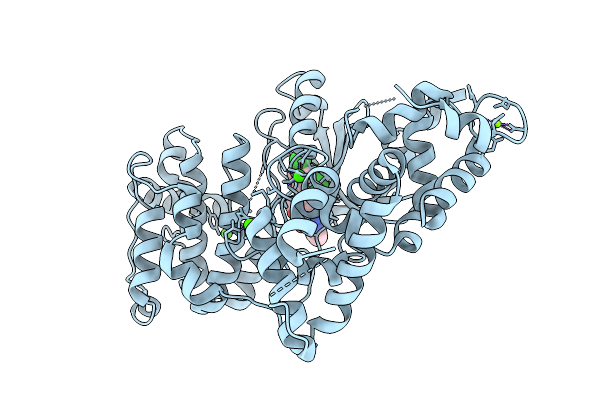 |
Crystal Structure Of Calcium-Dependent Protein Kinase 1 (Cdpk1) From Cryptosporidium Parvum In Complex With Inhibitor Win-3-115
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: CA, A1BLB, MG, CL |
 |
Crystal Structure Of Calcium-Dependent Protein Kinase 1 (Cdpk1) From Cryptosporidium Parvum In Complex With Inhibitor Bki-1606
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: A1BLC, CA |
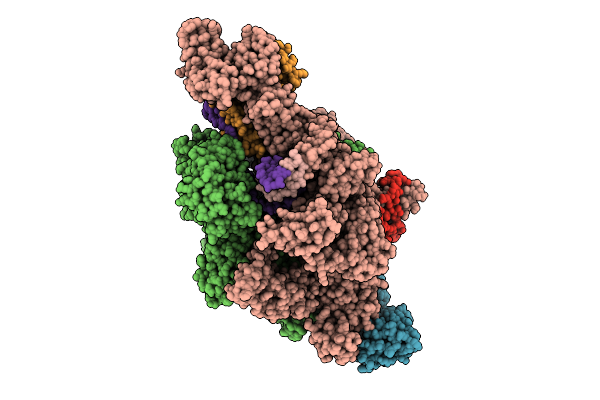 |
Organism: Thermus thermophilus hb8, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
Organism: Caloramator australicus
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: METAL BINDING PROTEIN Ligands: ZN, NA, FMT |
 |
Organism: Faecalibacterium prausnitzii l2-6
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE |
 |
Organism: Desulfovibrio desulfuricans atcc 27774
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: METAL BINDING PROTEIN Ligands: FE, A1CET, ACT, NA, 15P |
 |
Crystal Structure Of A Glyceraldehyde-3-Phosphate Dehydrogenase From Neisseria Gonorrhoeae In Complex With Nad And Glyceraldehyde-3-Phosphate
Organism: Neisseria gonorrhoeae nccp11945
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: OXIDOREDUCTASE Ligands: G3H, PEG, NAD, NA, PGE, CL, GOL, EPE |
 |
Crystal Structure Of Cryptosporidium Parvum N-Myristoyltransferase With Bound Myristoyl-Coa And Inhibitor 20045
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE/INHIBITOR Ligands: MYA, A1BHP, CL, PG4, 1PE, PGE |
 |
Crystal Structure Of Cryptosporidium Parvum N-Myristoyltransferase With Bound Myristoyl-Coa And Inhibitor 20057
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE/INHIBITOR Ligands: MYA, A1BHQ, CL, 1PE, PG4 |
 |
Crystal Structure Of Cryptosporidium Parvum N-Myristoyltransferase With Bound Myristoyl-Coa And Inhibitor 20084
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE/INHIBITOR Ligands: MYA, A1BHR, CL |

