Search Count: 1,996
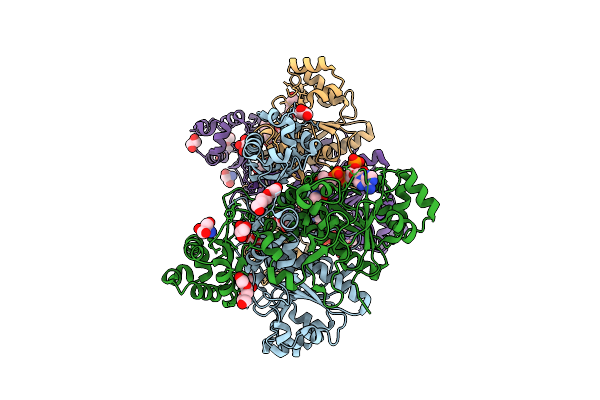 |
Succinyl-Coa:(R)-Benzylsuccinate Coa-Transferase (Bbsef), D178-Coa Adduct + Succinate (Weakly Occupied)
Organism: Aromatoleum aromaticum
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSFERASE Ligands: GOL, PGE, COA, TRS, SIN |
 |
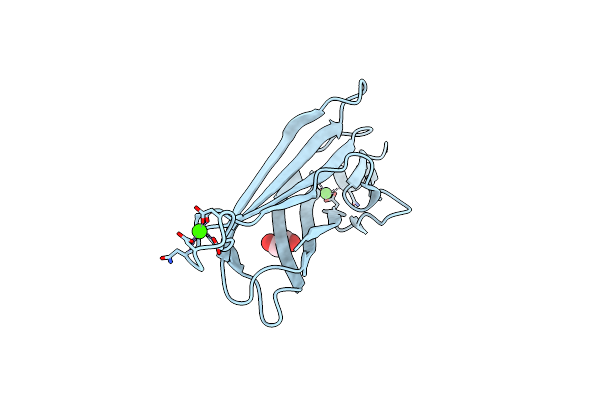
T450C Mutant Of Repeat Domain 2 From Clostridium Perfringens Adhesin Cpe0147 Without Intramolecular Ester Bond
Organism: Clostridium perfringens b str. atcc 3626
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: UNKNOWN FUNCTION Ligands: MG, CA |
 |
T450S Mutant Of Repeat Domain 2 From Clostridium Perfringens Adhesin Cpe0147 With Intramolecular Ester Bond
Organism: Clostridium perfringens b str. atcc 3626
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: UNKNOWN FUNCTION Ligands: EDO, CA |
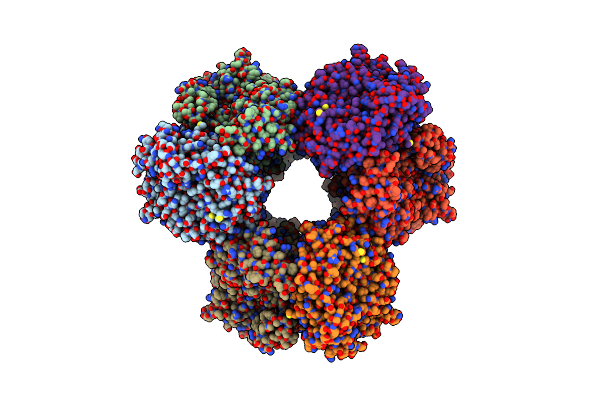 |
Enhancing The Synthesis Efficiency Of Galacto-Oligosaccharides Of A Beta-Galactosidase From Paenibacillus Barengoltzii By Engineering The Active And Distal Sites
Organism: Paenibacillus barengoltzii
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: HYDROLASE |
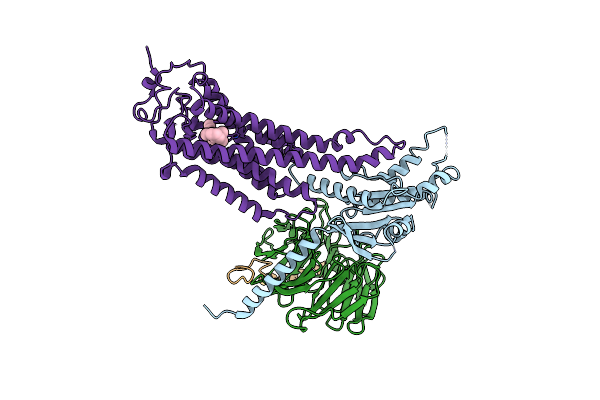 |
Organism: Homo sapiens, Hasarius adansoni, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: MEMBRANE PROTEIN Ligands: RET |
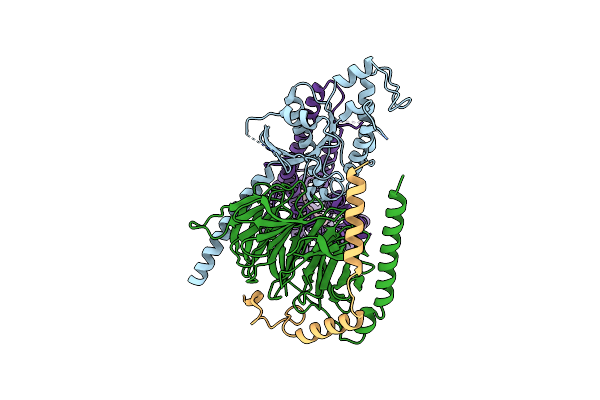 |
Cryo-Em Structure Of Jumping Spider Rhodopsin-1 Bound To A Giq Heterotrimer
Organism: Hasarius adansoni, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-23 Classification: MEMBRANE PROTEIN Ligands: A1H6M |
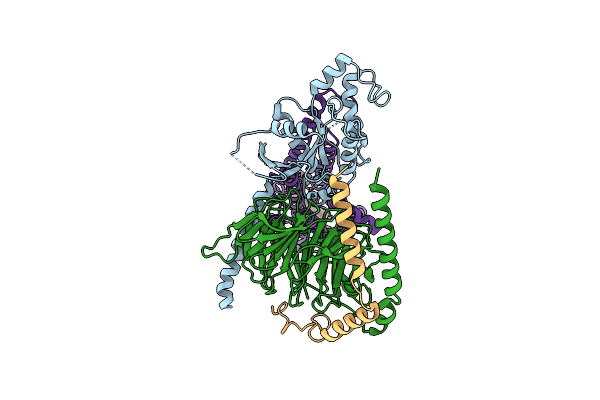 |
Cryo-Em Structure Of Jumping Spider Rhodopsin-1 Bound To A Giq Heterotrimer
Organism: Hasarius adansoni, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-23 Classification: MEMBRANE PROTEIN Ligands: A1H6M |
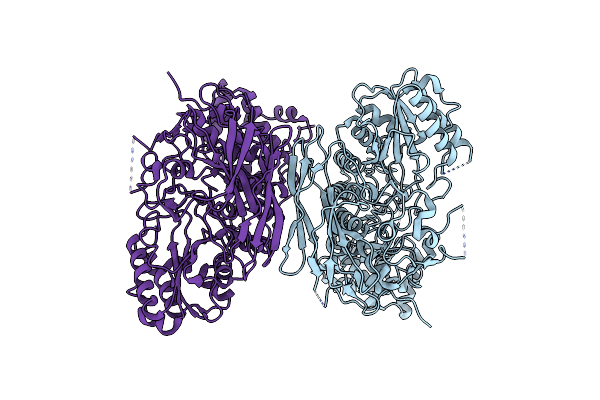 |
Organism: Prevotella copri
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2024-02-21 Classification: HYDROLASE |
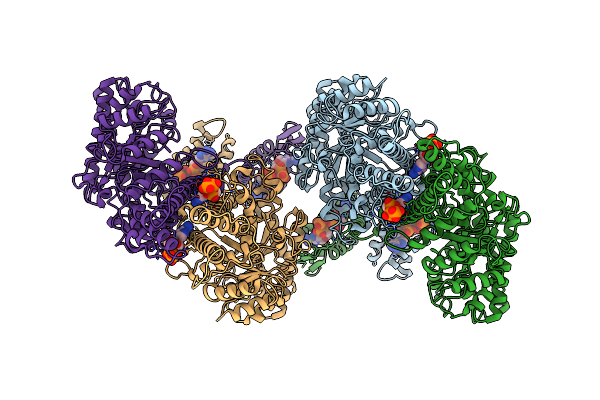 |
Cryo-Em Structure Of The Anaerobic Ribonucleotide Reductase From Prevotella Copri In Its Tetrameric State Produced In The Presence Of Datp And Ctp
Organism: Segatella copri
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: BIOSYNTHETIC PROTEIN Ligands: DTP, MG |
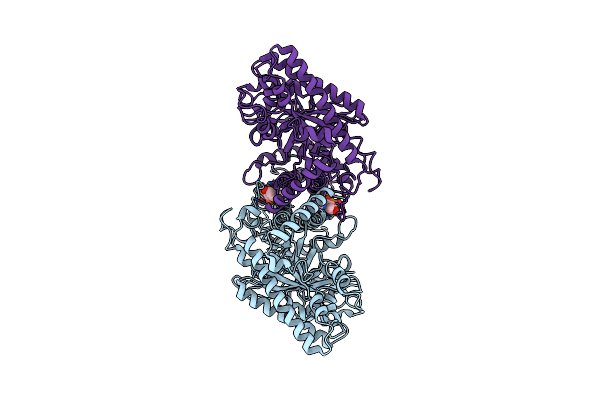 |
Cryo-Em Structure Of The Dimeric Form Of The Anaerobic Ribonucleotide Reductase From Prevotella Copri Produced In The Presence Of Datp And Ctp
Organism: Segatella copri
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: BIOSYNTHETIC PROTEIN Ligands: DTP, MG |
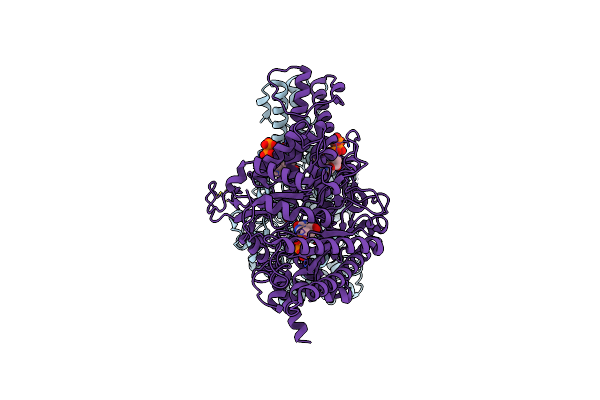 |
Cryo-Em Structure Of The Anaerobic Ribonucleotide Reductase From Prevotella Copri In Its Dimeric, Atp/Dttp/Gtp-Bound State
Organism: Segatella copri
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: BIOSYNTHETIC PROTEIN Ligands: TTP, MG, GTP, ZN |
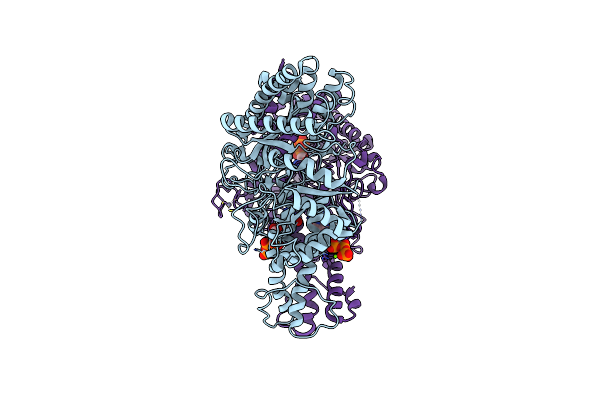 |
Cryo-Em Structure Of The Anaerobic Ribonucleotide Reductase From Prevotella Copri In Its Dimeric, Dgtp/Atp-Bound State
Organism: Segatella copri
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: BIOSYNTHETIC PROTEIN Ligands: DGT, MG, ATP, ZN |
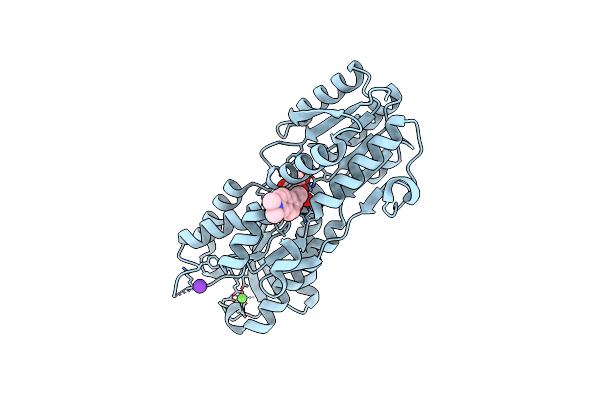 |
Thermoanaerobacter Thermosaccharolyticum Periplasmic Glucose-Binding Protein Glucose Complex: Badan Conjugate Attached At F17C
Organism: Thermoanaerobacterium thermosaccharolyticum
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2023-08-30 Classification: SUGAR BINDING PROTEIN Ligands: BGC, CA, K, YDQ, CL |
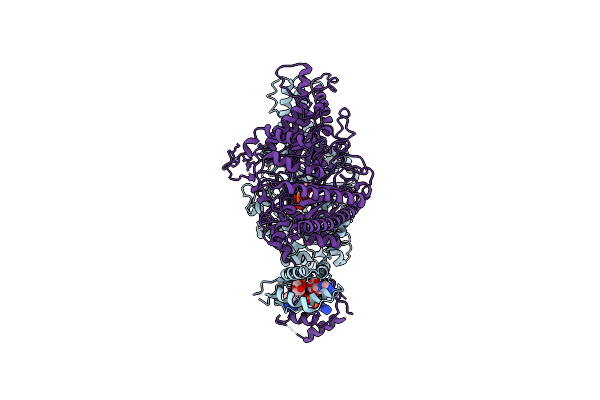 |
Cryo-Em Structure Of The Anaerobic Ribonucleotide Reductase From Prevotella Copri In Its Dimeric, Atp/Ctp-Bound State
Organism: Prevotella copri
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: BIOSYNTHETIC PROTEIN Ligands: ATP, CTP, ZN |
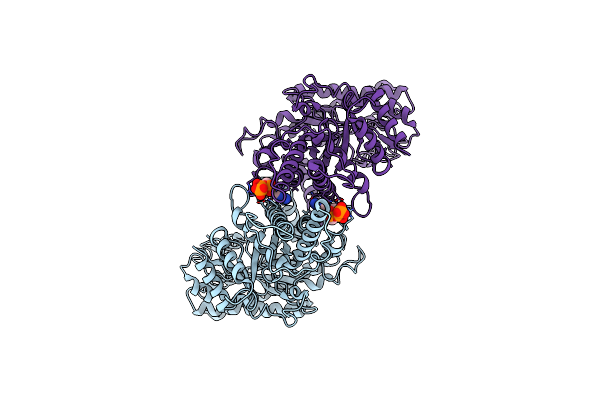 |
Cryo-Em Structure Of The Anaerobic Ribonucleotide Reductase From Prevotella Copri In Its Dimeric, Datp-Bound State
Organism: Segatella copri
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: BIOSYNTHETIC PROTEIN Ligands: DTP, MG |
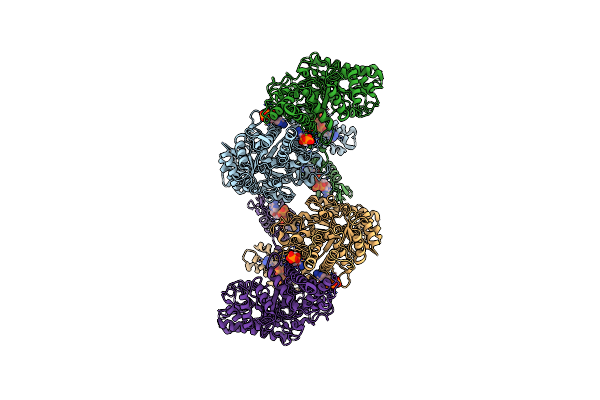 |
Cryo-Em Structure Of The Anaerobic Ribonucleotide Reductase From Prevotella Copri In Its Tetrameric, Datp-Bound State
Organism: Segatella copri
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: OXIDOREDUCTASE Ligands: DTP, MG |
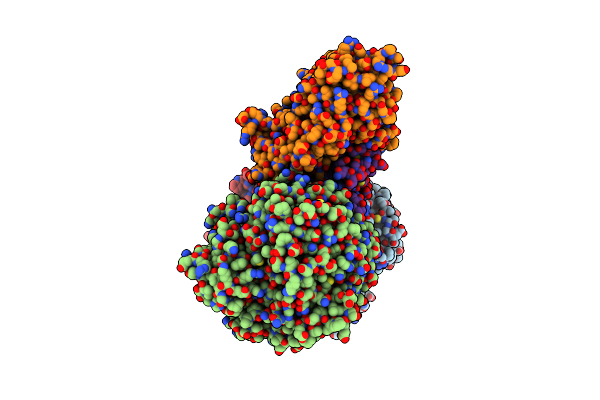 |
Cryoem Structure Of A Tungsten-Containing Aldehyde Oxidoreductase From Aromatoleum Aromaticum
Organism: Aromatoleum aromaticum
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: OXIDOREDUCTASE Ligands: SF4, T7R, MG, BEZ, FAD |
 |
Organism: Cecembia lonarensis
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2023-03-08 Classification: TRANSFERASE |
 |
Organism: Cecembia lonarensis lw9
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2023-01-18 Classification: HYDROLASE |
 |
Organism: Cecembia lonarensis lw9
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2023-01-18 Classification: HYDROLASE |

