Search Count: 221
 |
Organism: Guillardia theta ccmp2712
Method: ELECTRON MICROSCOPY Resolution:2.86 Å Release Date: 2024-09-04 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
Organism: Guillardia theta ccmp2712
Method: ELECTRON MICROSCOPY Resolution:2.73 Å Release Date: 2024-09-04 Classification: MEMBRANE PROTEIN Ligands: RET, PC1 |
 |
Crystal Structure Of Group 1 Oligosaccharide-Releasing Beta-N-Acetylgalactosaminidase Ngaca From Cohnella Abietis, Apo 1 Form
Organism: Cohnella abietis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: MG |
 |
Crystal Structure Of Group 1 Oligosaccharide-Releasing Beta-N-Acetylgalactosaminidase Ngaca From Cohnella Abietis, Apo 2 Form
Organism: Cohnella abietis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: MG |
 |
Organism: Cutibacterium acnes hl110pa3
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-12-13 Classification: LYASE Ligands: GOL, PO4 |
 |
Crystal Structure Of Y281F Mutant Of Hyaluronate Lyase B From Cutibacterium Acnes
Organism: Cutibacterium acnes hl110pa3
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-12-13 Classification: LYASE |
 |
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2023-04-05 Classification: BIOSYNTHETIC PROTEIN Ligands: NAG, CA, BEZ, EDO, PEG, GOL, MAN |
 |
Organism: Guillardia theta ccmp2712
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-08-07 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Organism: Guillardia theta ccmp2712
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-08-07 Classification: OXIDOREDUCTASE Ligands: DBV, SO4, 1PE |
 |
Organism: Guillardia theta ccmp2712
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-01-16 Classification: MEMBRANE PROTEIN Ligands: OLC, GOL |
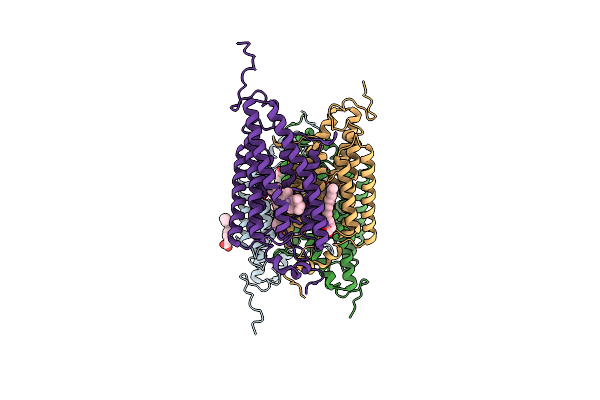 |
Organism: Guillardia theta ccmp2712
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-09-05 Classification: MEMBRANE PROTEIN Ligands: RET, OLA |
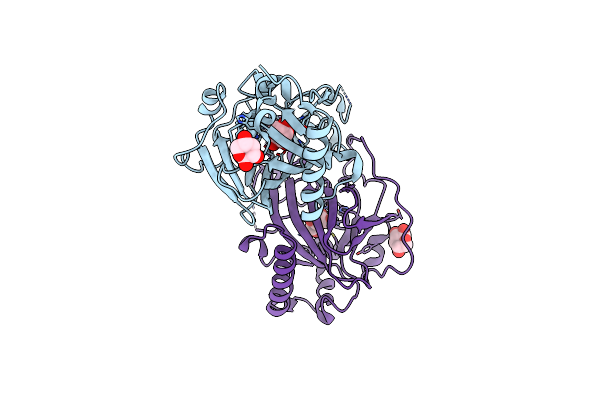 |
X-Ray Structure Of Cellulomonas Parahominis L-Ribose Isomerase With L-Ribose
Organism: Cellulomonas parahominis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2015-04-29 Classification: ISOMERASE Ligands: MN, 0MK, ROR |
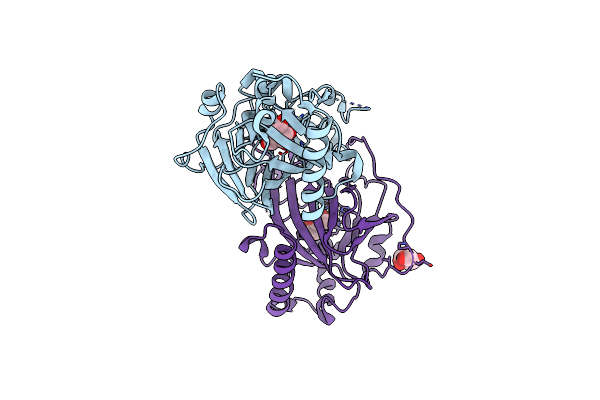 |
X-Ray Structures Of Cellulomonas Parahominis L-Ribose Isomerase With L-Psicose
Organism: Cellulomonas parahominis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-04-29 Classification: ISOMERASE Ligands: MN, SF6, LPK, SF9 |
 |
X-Ray Structures Of Cellulomonas Parahominis L-Ribose Isomerase With No Ligand
Organism: Cellulomonas parahominis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-04-29 Classification: ISOMERASE Ligands: MN |
 |
X-Ray Structures Of Cellulomonas Parahominis L-Ribose Isomerase With L-Allose
Organism: Cellulomonas parahominis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2015-04-29 Classification: ISOMERASE Ligands: MN, 3BU, WOO |
 |
Organism: Guillardia theta ccmp2712
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2014-08-13 Classification: LYASE Ligands: HEZ |
 |
Crystallographic Analysis Of Transition State Mimics Bound To Penicillopepsin: Phosphorous-Containing Peptide Analogues
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1994-05-31 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: IVV, MAN, HSY, SO4, DMF |
 |
Crystallographic Analysis Of A Pepstatin Analogue Binding To The Aspartyl Proteinase Penicillopepsin At 1.8 Angstroms Resolution
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1994-01-31 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: MAN |
 |
Crystallographic Analysis Of A Pepstatin Analogue Binding To The Aspartyl Proteinase Penicillopepsin At 1.8 Angstroms Resolution
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1994-01-31 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: MAN |
 |
Crystallographic Analysis Of Transition State Mimics Bound To Penicillopepsin: Difluorostatine-And Difluorostatone-Containing Peptides
Organism: Penicillium janthinellum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1994-01-31 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: MAN, XYS, SO4, DMF |

