Search Count: 96
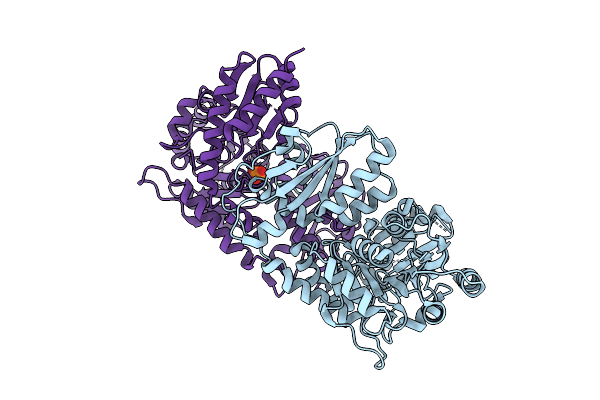 |
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: LYASE Ligands: PO4, CL |
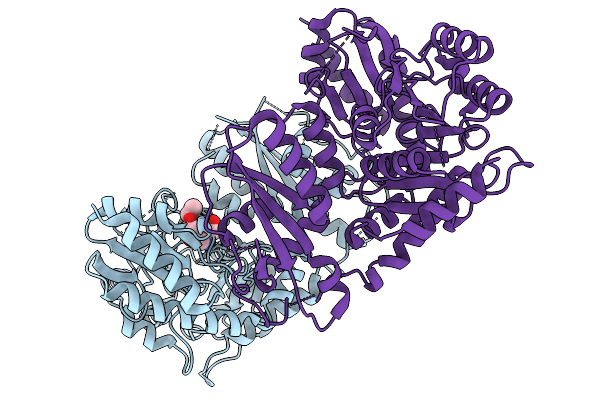 |
Gamma-Lyase Cndf In Complex With Pyridoxal 5'-Phosphate, L-Homoserine And Ethyl Acetoacetate
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: LYASE Ligands: HSE, EAC |
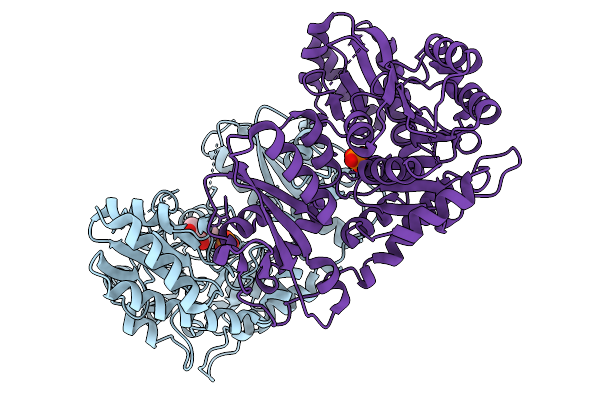 |
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: LYASE Ligands: PO4, EAC |
 |
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2023-11-29 Classification: HYDROLASE Ligands: RBH, XYS, YLL |
 |
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2023-11-29 Classification: HYDROLASE |
 |
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2023-11-22 Classification: HYDROLASE |
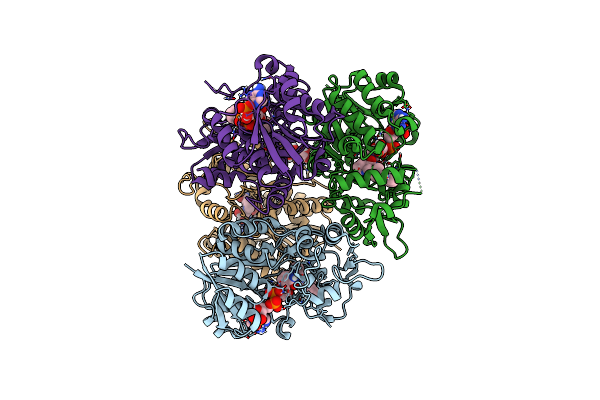 |
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-02-22 Classification: OXIDOREDUCTASE Ligands: NAP, N3L, EDO |
 |
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-06-16 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD |
 |
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2021-06-16 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, WU4, EDO, CL |
 |
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2020-08-19 Classification: HYDROLASE |
 |
Crystal Structure Of Cjagd31B (Alpha-Transglucosylase From Glycoside Hydrolase Family 31) In Complex With Alpha Cyclophellitol Cyclosulfate Probe Me647
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-08-09 Classification: HYDROLASE Ligands: SO4, PG4, 93Z, OXL |
 |
Crystal Structure Of D412N Nucleophile Mutant Cjagd31B (Alpha-Transglucosylase From Glycoside Hydrolase Family 31) In Complex With Unreacted Alpha Cyclophellitol Cyclosulfate Probe Me647
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2017-08-09 Classification: HYDROLASE Ligands: SO4, EDO, PG4, 94E, OXL |
 |
Crystal Structure Of D412N Nucleophile Mutant Cjagd31B (Alpha-Transglucosylase From Glycoside Hydrolase Family 31) In Complex With Alpha Cyclophellitol Aziridine Probe Cf021
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-08-09 Classification: HYDROLASE Ligands: SO4, PG4, EDO, 94B, OXL |
 |
Organism: Burkholderia cenocepacia bc7
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-03-22 Classification: UNKNOWN FUNCTION Ligands: OTP, SO4, TRS |
 |
Organism: Microcystis aeruginosa mrc
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2016-09-21 Classification: LIGASE |
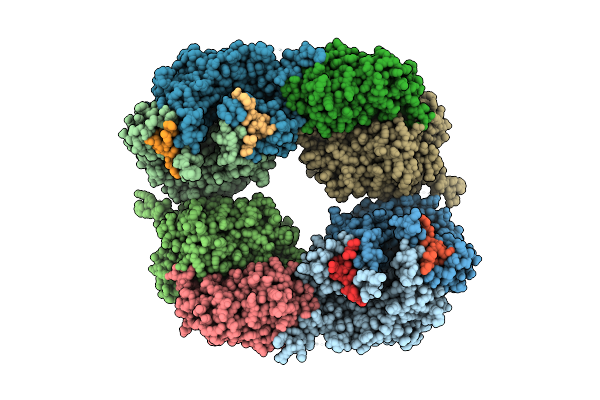 |
Crystal Structure Of Macrocyclase Mdnc Bound With Precursor Peptide Mdna From Microcystis Aeruginosa Mrc
Organism: Microcystis aeruginosa mrc
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2016-09-21 Classification: LIGASE |
 |
Structural And Functional Analysis Of A Lytic Polysaccharide Monooxygenase Important For Efficient Utilization Of Chitin In Cellvibrio Japonicus
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-02-17 Classification: SUGAR BINDING PROTEIN Ligands: CU |
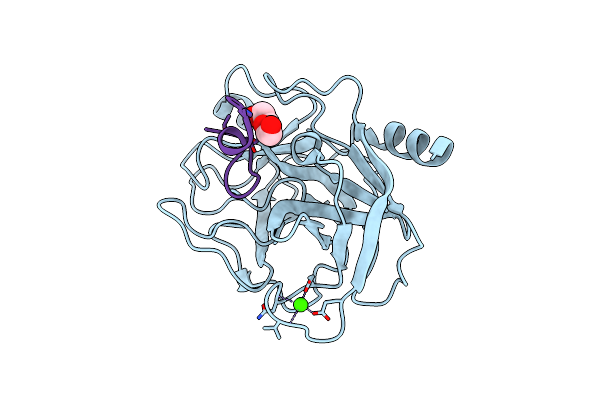 |
Organism: Microcystis aeruginosa mrc, Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2014-04-02 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA, PEG |
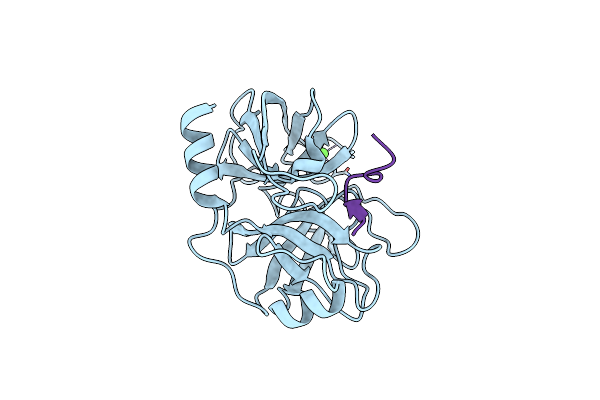 |
Organism: Microcystis aeruginosa mrc, Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2014-04-02 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA |
 |
Crystal Structure Of Apo Agd31B, Alpha-Transglucosylase In Glycoside Hydrolase Family 31
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-11-14 Classification: HYDROLASE Ligands: SO4, OXL, EDO, PG4 |

