Search Count: 630
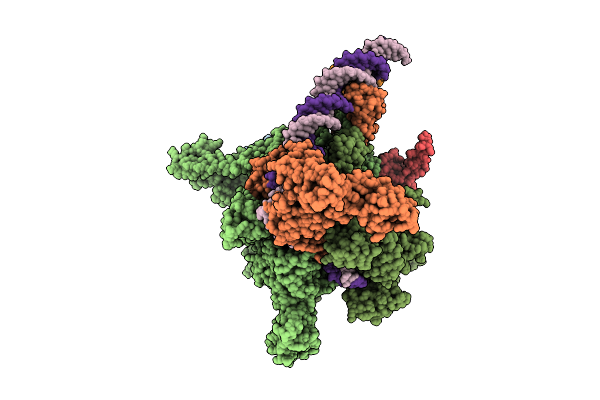 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Holoenzyme Bound To An Ompu Promoter Dna Fragment
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
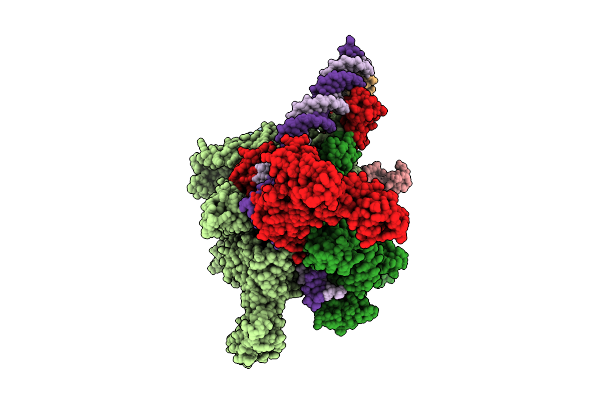 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Holoenzyme Bound To An Ompu Promoter Dna Fragment And 5-Mer Rna
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
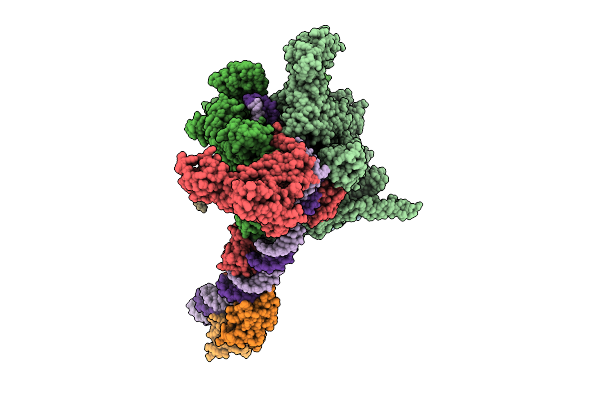 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Transcription Activation Complex With Toxr Transcription Factor And Ompu Promoter Dna
Organism: Vibrio cholerae o395, Vibrio cholerae o1 biovar el tor str. n16961
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
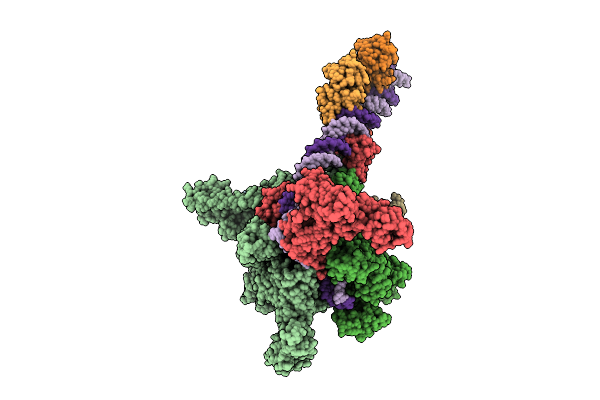 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Transcription Activation Complex With Tcpp Transcription Factor And A Toxt Promoter Dna Fragment
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
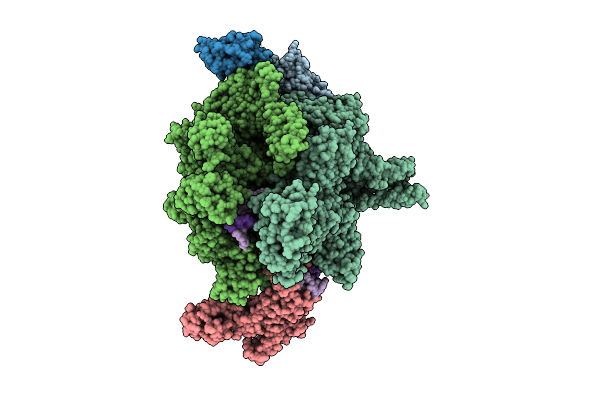 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Transcription Activation Complex With Toxr And Tcpp Transcription Factors And A Toxt Promoter Dna Fragment
Organism: Vibrio cholerae o395, Vibrio cholerae o1 biovar el tor str. n16961
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
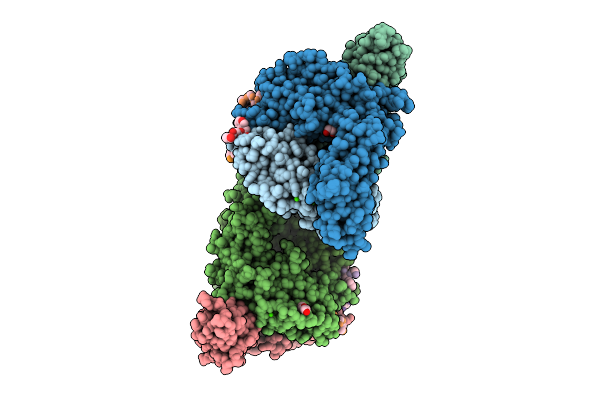 |
Vibrio Cholerae Protein Frha Peptid-Binding Domain And Adjacent Split Domain (S1127-F1439) In Complex With Peptide Agwtd X-Ray Crystallography Structure
Organism: Vibrio cholerae o395, Vibrio cholerae
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: METAL BINDING PROTEIN Ligands: CA, PEG, EDO |
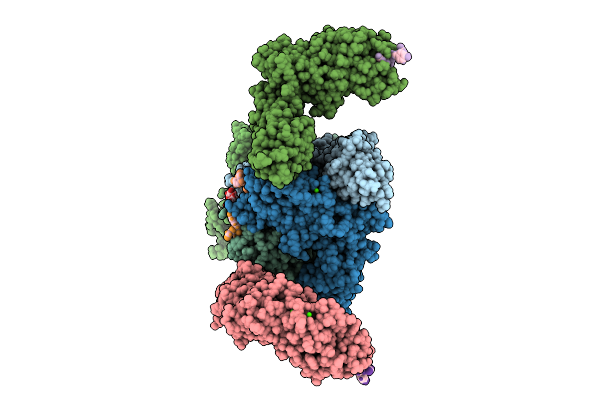 |
Vibrio Cholerae Protein Frha Peptid-Binding Domain And Adjacent Split Domain (S1127-F1439) In Complex With Peptide Agytd X-Ray Crystallography Structure
Organism: Vibrio cholerae o395, Vibrio cholerae
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: METAL BINDING PROTEIN Ligands: CA |
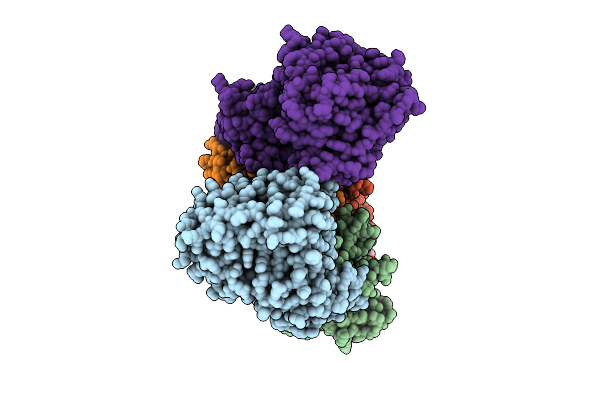 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, In The Absence Of Na+, Middle State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, CA, LMT, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrc-T225Y Mutant From Vibrio Cholerae
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, LMT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrc-T225Y Mutant From Vibrio Cholerae Reduced By Nadh
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, LMT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-T236Y Mutant From Vibrio Cholerae
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: RBF, LMT, FMN, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-T236Y Mutant From Vibrio Cholerae Reduced By Nadh
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: RBF, LMT, CA, FMN, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, With Bound Korormicin A
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, IQT, LMT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, In The Absence Of Na+, Upper State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, LMT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, In The Absence Of Na+, Down State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-G141A Mutant From Vibrio Cholerae Reduced By Nadh, With Bound Korormicin A, Stable State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, IQT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-G141A Mutant From Vibrio Cholerae Reduced By Nadh, With Bound Korormicin A, Shifted State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, PEE, LMT, IQT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, With Bound Aurachin D-42
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, 0NI, PEE, LMT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, CA, FES, FAD, NAI |
 |
Organism: Penicillium
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: BIOSYNTHETIC PROTEIN Ligands: FE |

