Search Count: 29
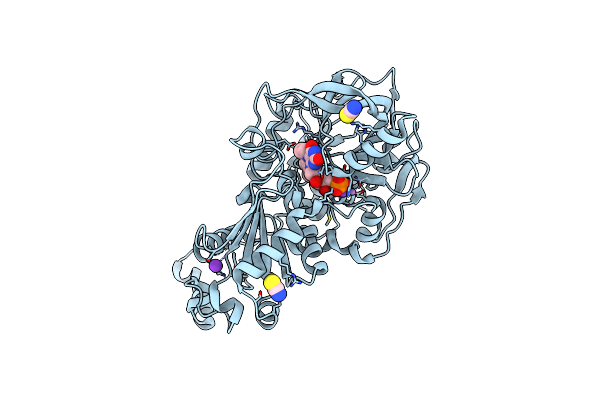 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) C122-S261C (Db3) Variant In Complex With Prenylated Flavin
Organism: Aspergillus niger cbs 513.88
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2023-09-06 Classification: LYASE Ligands: 4LU, MN, K, SCN |
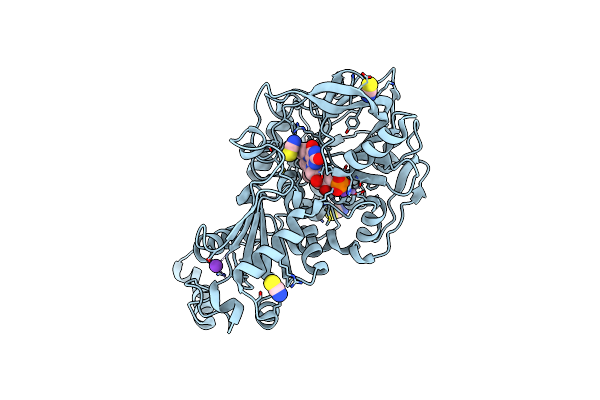 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) V186C-A296C (Db4) Variant In Complex With Prenylated Flavin
Organism: Aspergillus niger cbs 513.88
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-09-06 Classification: LYASE Ligands: 4LU, MN, K, SCN |
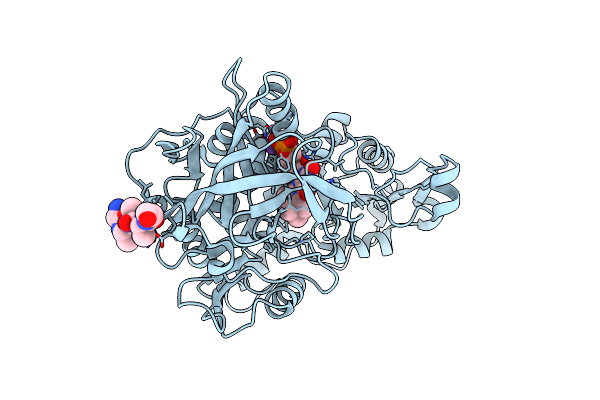 |
Crystal Structure Of Phenazine 1-Carboxylic Acid Decarboxylase From Mycobacterium Fortuitum
Organism: Mycolicibacterium fortuitum, Unidentified
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2022-08-24 Classification: LYASE Ligands: MN, 4LU, NA |
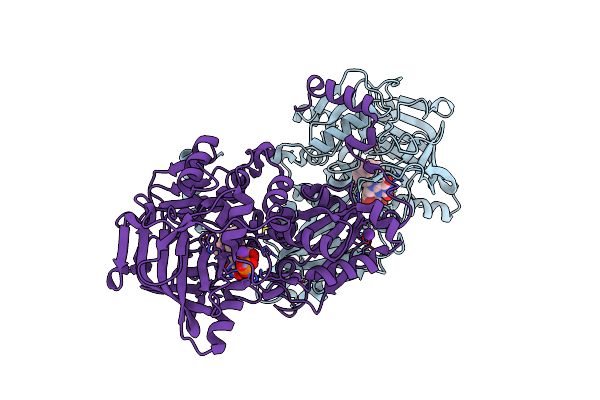 |
Organism: Hypocrea atroviridis (strain atcc 20476 / imi 206040)
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2021-08-04 Classification: LYASE Ligands: 4LU, K, MN |
 |
Organism: Aspergillus niger (strain cbs 513.88 / fgsc a1513)
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2021-08-04 Classification: LYASE Ligands: K, 4LU, MN |
 |
Structure Of The Reversible Pyrrole-2-Carboxylic Acid Decarboxylase Pa0254/Huda
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-04-07 Classification: LIGASE Ligands: MN, K, 4LU, IMD |
 |
Crystal Structure Of Ubix-Like Fmn Prenyltransferase Af1214 From Archaeoglobus Fulgidus, Prenylated-Fmn Complex
Organism: Archaeoglobus fulgidus (strain atcc 49558 / vc-16 / dsm 4304 / jcm 9628 / nbrc 100126)
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2020-02-26 Classification: LYASE Ligands: 4LU, PO4 |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) In Complex With Prfmn (Purified In The Radical Form) And Phenylpropiolic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2019-08-28 Classification: LYASE Ligands: 4LU, MN, K, JQ8 |
 |
Aspergillus Niger Ferric Acid Decarboxylase (Fdc) L439G Variant In Complex With Prfmn (Purified In The Radical Form) And Phenylpropiolic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2019-08-28 Classification: LYASE Ligands: 4LU, MN, K, JQ8 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2019-06-05 Classification: TRANSFERASE Ligands: 4LU, PO4, NA |
 |
Structure Of An Evolved Dimeric Form Of The Ubid-Class Enzyme Hmff From Pelotomaculum Thermopropionicum In Complex With Prfmn
Organism: Pelotomaculum thermopropionicum (strain dsm 13744 / jcm 10971 / si)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-02-27 Classification: FLAVOPROTEIN Ligands: K, MN, 4LU, CA |
 |
Structure Of Wild Type A. Niger Fdc1 Purified In The Dark With Prfmn In The Iminium Form
Organism: Aspergillus niger (strain cbs 513.88 / fgsc a1513)
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2017-12-27 Classification: LYASE Ligands: 4LU, MN, K |
 |
Structure Of Wild Type A. Niger Fdc1 That Has Been Illuminated With Uv Light, With Prfmn In The Iminium And Ketimine Form
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2017-12-20 Classification: LYASE Ligands: 4LU, MN, K, FZZ |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2017-12-20 Classification: LYASE Ligands: MN, K, 4LU |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2017-12-20 Classification: LYASE Ligands: 4LU, MN, K, SCN |
 |
Organism: Aspergillus niger (strain cbs 513.88 / fgsc a1513)
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2017-12-20 Classification: LYASE Ligands: 4LU, MN, K, F5C |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2017-12-20 Classification: LYASE Ligands: 4LU, MN, K |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2017-12-20 Classification: LYASE Ligands: 4LU, MN, K |
 |
Crystal Structure Of E. Cloacae 3,4-Dihydroxybenzoic Acid Decarboxylase (Aroy) Reconstituted With Prfmn
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2017-09-13 Classification: LYASE Ligands: MN, NA, GOL, 4LU |
 |
Crystal Structure Of N-Terminally Tagged Ubid From E. Coli Reconstituted With Prfmn Cofactor
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-01-11 Classification: LYASE Ligands: MN, 4LU, NA, SO4 |

