Search Count: 6
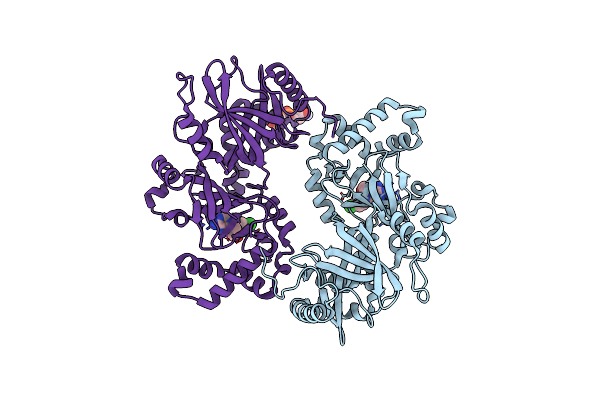 |
Crystal Structure Of C.Difficile Ribosyltransferase Cdta In Complex With Pcl-Phenylthiodadmeimma
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2021-10-13 Classification: TOXIN, TRANSFERASE Ligands: 4CT, AR6 |
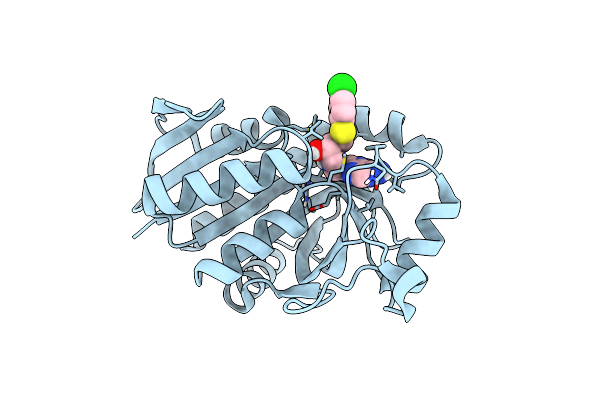 |
Organism: Helicobacter pylori
Method: NEUTRON DIFFRACTION, X-RAY DIFFRACTION Resolution:2.60 Å, 2.25 Å Release Date: 2016-11-16 Classification: HYDROLASE Ligands: 4CT, DOD |
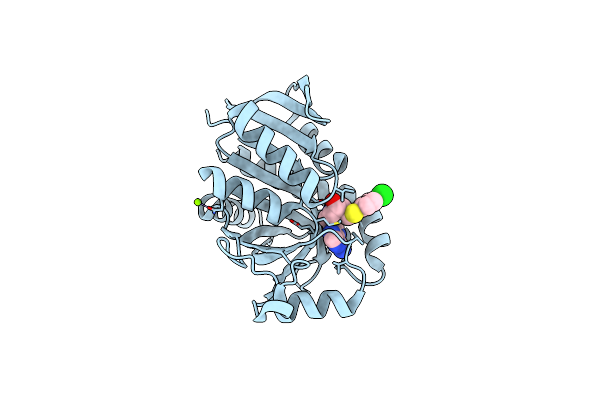 |
1.4 A Resolution Structure Of Helicobacter Pylori Mtan In Complexed With P-Clph-Dadme-Imma
Organism: Helicobacter pylori (strain j99 / atcc 700824)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-11-16 Classification: HYDROLASE Ligands: 4CT, MG |
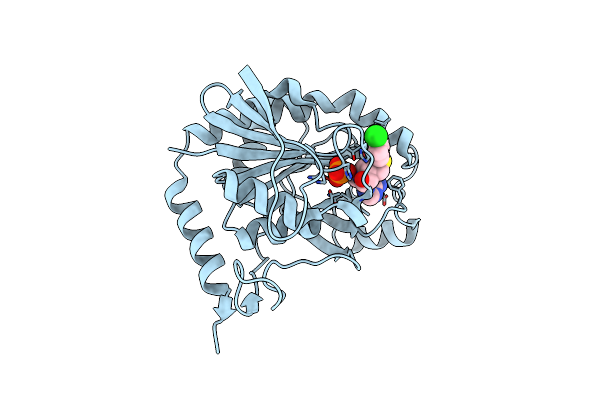 |
Crystal Structure Of Human 5'-Deoxy-5'-Methyladenosine Phosphorylase In Complex With Pcl-Phenylthiodadmeimma
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2011-09-28 Classification: TRANSFERASE Ligands: 4CT, PO4 |
 |
Crystal Structure Of Human 5'-Deoxy-5'-Methyladenosine Phosphorylase In Complex With Pcl-Phenylthiodadmeimma
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-09-28 Classification: TRANSFERASE Ligands: 4CT |
 |
Crystal Structure Of E. Coli Mta/Sah Nucleosidase In Complex With (4-Chlorophenyl)Thio-Dadme-Imma
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2010-08-11 Classification: HYDROLASE Ligands: 4CT, GOL, IPA |

