Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
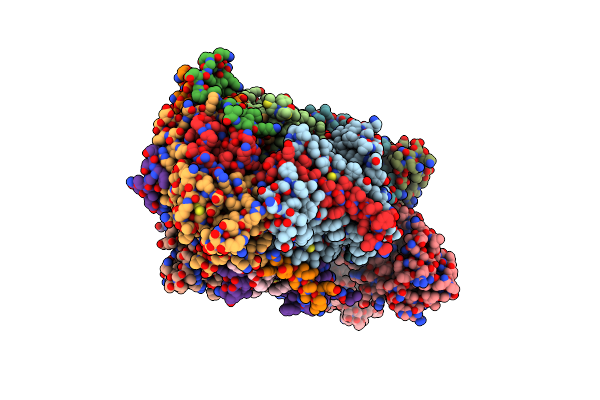 |
Late Assembly Intermediate Of The Proximal Proton Pumping Module Of Complex I With Assembly Factors Ndufaf1 And Cia84
Organism: Yarrowia lipolytica
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: ELECTRON TRANSPORT Ligands: CDL, PLC, CPL, 3PE, T7X |
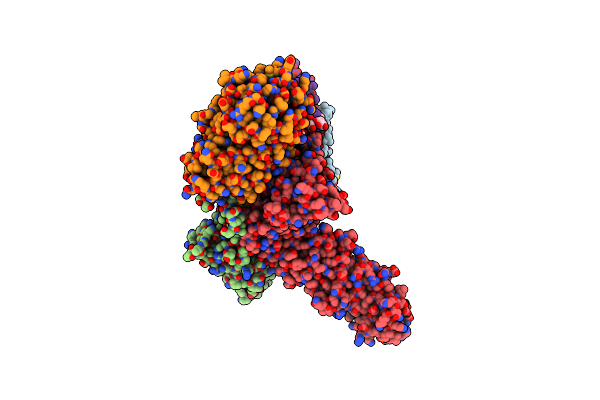 |
Organism: Yarrowia lipolytica
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: ELECTRON TRANSPORT Ligands: CDL, LMN, PLC |
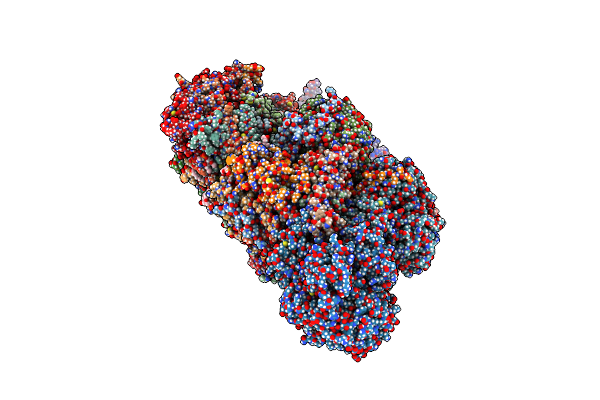 |
A 3.7-Angstrom Structure Of Yarrowia Lipolytica Complex I With An R121M Mutation In Nucm.
Organism: Yarrowia lipolytica
Method: ELECTRON MICROSCOPY Release Date: 2021-03-10 Classification: MEMBRANE PROTEIN Ligands: PLC, FES, FMN, SF4, 3PE, CDL, NDP, ZN, EHZ, LMT |
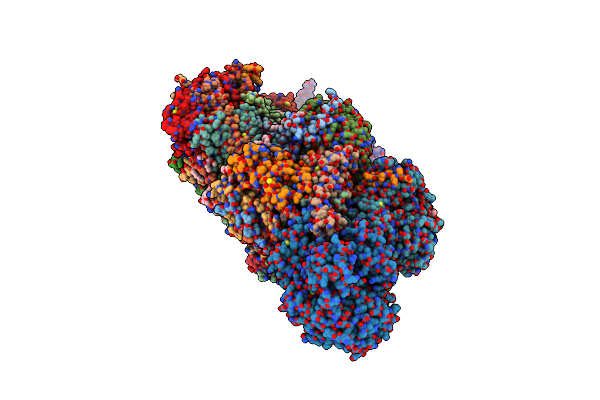 |
Organism: Yarrowia lipolytica, Yarrowia lipolytica (strain clib 122 / e 150)
Method: ELECTRON MICROSCOPY Release Date: 2020-08-12 Classification: MEMBRANE PROTEIN Ligands: 3PE, SF4, LMT, PLC, FES, FMN, CDL, NDP, ZN, EHZ |
 |
Focused Refinement Cryo-Em Structure Of The Yeast Mitochondrial Complex I Sub-Stoichiometric Sulfur Transferase Subunit
Organism: Yarrowia lipolytica
Method: ELECTRON MICROSCOPY Release Date: 2020-08-12 Classification: TRANSFERASE |
 |
Organism: Yarrowia lipolytica
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2020-05-20 Classification: LIPID BINDING PROTEIN Ligands: PO4 |
 |
Organism: Enterococcus faecium tx0133a04
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-07-03 Classification: TRANSFERASE Ligands: GOL |
 |
Organism: Escherichia coli, Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2013-12-11 Classification: REPLICATION |
 |
Organism: Yarrowia lipolytica
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-03-13 Classification: HYDROLASE |
 |
Organism: Hahella chejuensis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2012-10-17 Classification: HYDROLASE Ligands: PEG, GOL, PO4, ACT, EDO, CA, NA |
 |
Structure Of Candida Albicans Kar3 Motor Domain Fused To Maltose-Binding Protein
Organism: Escherichia coli, Candida albicans
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2012-10-10 Classification: MOTOR PROTEIN Ligands: MG, ADP, EDO |
 |
Crystal Structure Of Lip2 Lipase From Yarrowia Lipolytica At 1.7 A Resolution
Organism: Yarrowia lipolytica
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-11-24 Classification: HYDROLASE Ligands: NAG, MPD, MRD, K |
 |
Organism: Hahella chejuensis
Method: SOLUTION NMR Release Date: 2010-11-10 Classification: CALCIUM-BINDING PROTEIN |
 |
Crystal Structure Of Abc-Type Sugar Transport System, Periplasmic Component From Hahella Chejuensis
Organism: Hahella chejuensis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-12-15 Classification: TRANSPORT PROTEIN Ligands: BDR |
 |
Organism: Hahella chejuensis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-11-10 Classification: SIGNALING PROTEIN |
 |
 |
 |
 |
 |