Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Kitasatospora mediocidica kctc 9733
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-11-20 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Crystal Structure Of Ptmb In Complex With Cyclo-(L-Trp-L-Trp) And Hypoxanthine
Organism: Kitasatospora mediocidica kctc 9733
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-11-20 Classification: OXIDOREDUCTASE Ligands: UYM, HPA, HEM |
 |
Organism: Kitasatospora mediocidica kctc 9733
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-11-20 Classification: OXIDOREDUCTASE Ligands: ADE, HEM, UYM |
 |
Organism: Kitasatospora mediocidica kctc 9733
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2024-11-20 Classification: OXIDOREDUCTASE Ligands: HEM, ADE |
 |
Organism: Kitasatospora mediocidica kctc 9733
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-11-20 Classification: OXIDOREDUCTASE Ligands: UYM, GUN, HEM |
 |
Organism: Streptococcus pyogenes m49 591
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-03-16 Classification: OXIDOREDUCTASE Ligands: SO4, GOL |
 |
Organism: Streptococcus pyogenes m49 591
Method: X-RAY DIFFRACTION Release Date: 2022-03-16 Classification: OXIDOREDUCTASE Ligands: BME, SO4, GOL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Bat coronavirus rp3/2004, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-02-23 Classification: VIRAL PROTEIN Ligands: NAG |
 |
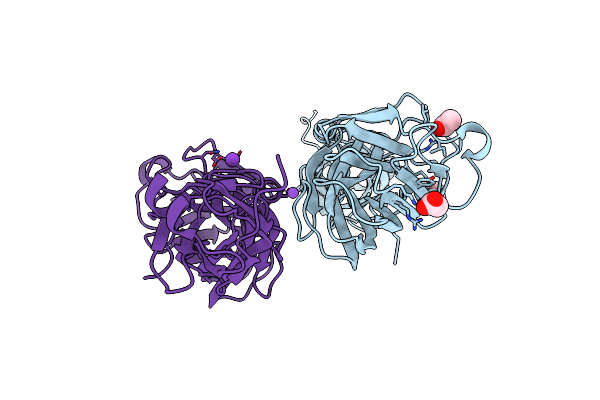
Crystal Structure Of Streptococcus Pyogenes Endo-Beta-N-Acetylglucosaminidase (Endos2)
Organism: Streptococcus pyogenes m49 591
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2019-02-06 Classification: HYDROLASE Ligands: CA |
 |
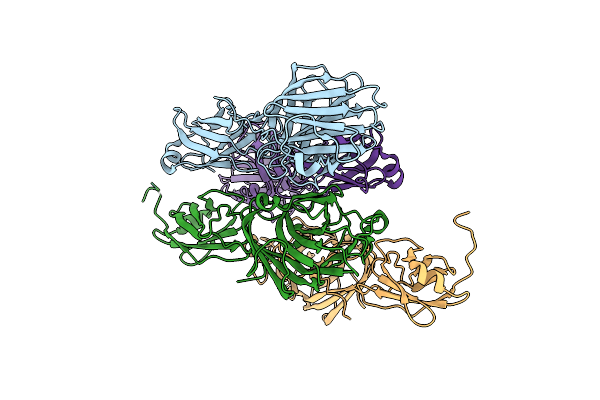
Crystal Structure Of The Adhesin Domain Of Epf From Streptococcus Pyogenes In P212121
Organism: Streptococcus pyogenes
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2012-09-19 Classification: CELL ADHESION Ligands: GOL, ACT, NA, K |
 |
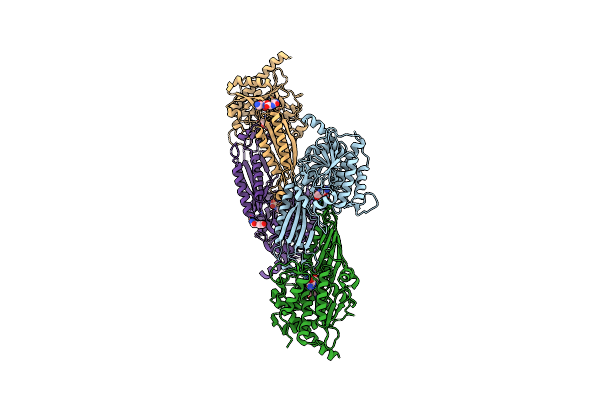
Crystal Structure Of The Adhesin Domain Of Epf From Streptococcus Pyogenes In P21
Organism: Streptococcus pyogenes
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-09-19 Classification: CELL ADHESION |
 |
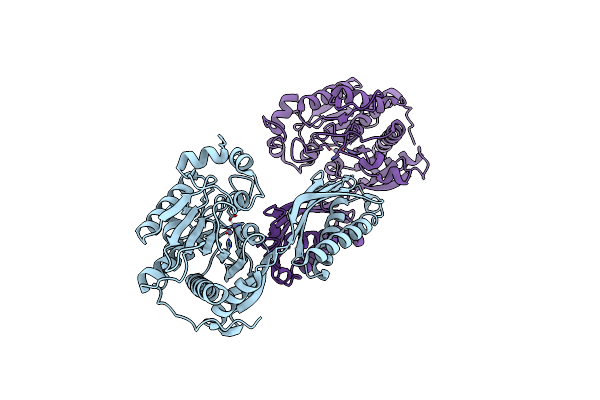
Crystal Structure Of Beta-Alanine Synthase From Saccharomyces Kluyveri In Complex With A Gly-Gly Peptide
Organism: Saccharomyces kluyveri
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2008-05-13 Classification: HYDROLASE Ligands: GLY, ZN |
 |
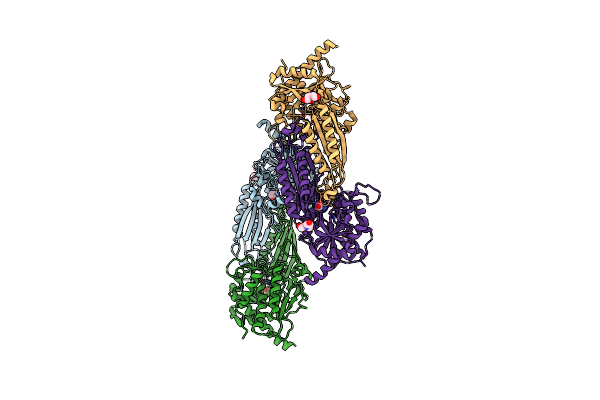
Crystal Structure Of Mutant E159A Of Beta-Alanine Synthase From Saccharomyces Kluyveri
Organism: Saccharomyces kluyveri
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-10-02 Classification: HYDROLASE Ligands: ZN |
 |
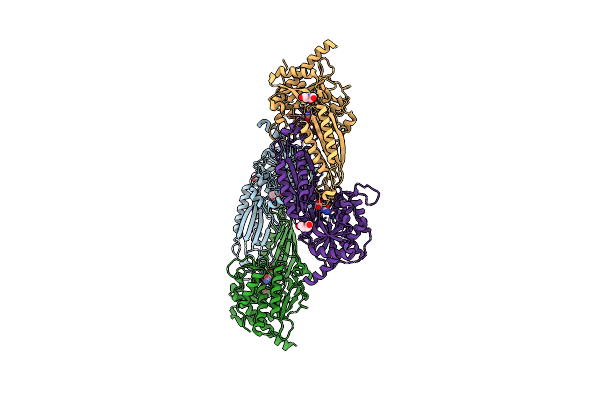
Crystal Structure Of Beta-Alanine Synthase From Saccharomyces Kluyveri In Complex With The Product Beta-Alanine
Organism: Saccharomyces kluyveri
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-10-02 Classification: HYDROLASE Ligands: ZN, BAL, BCN |
 |
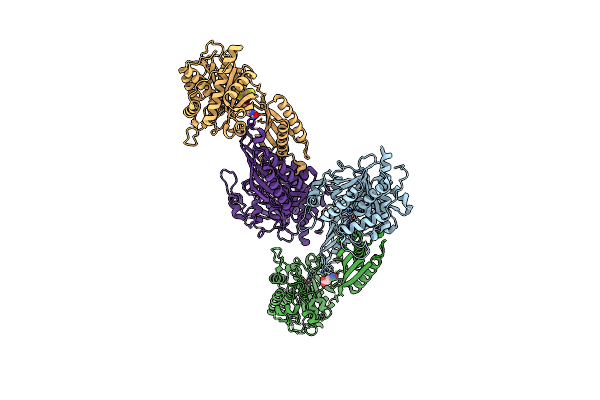
Crystal Structure Of Mutant E159A Of Beta-Alanine Synthase From Saccharomyces Kluyveri In Complex With Its Substrate N-Carbamyl-Beta- Alanine
Organism: Saccharomyces kluyveri
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-10-02 Classification: HYDROLASE Ligands: ZN, URP, BCN |
 |
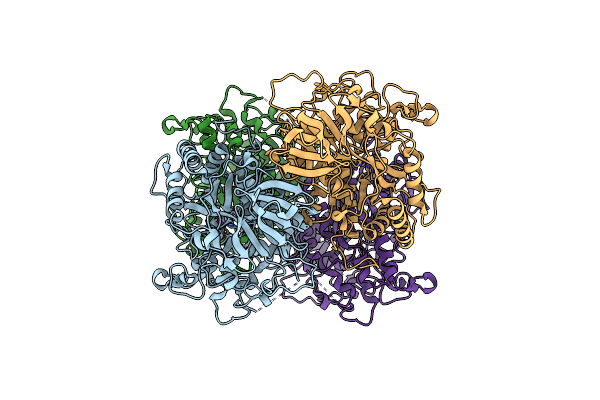
Crystal Structure Of Mutant R322A Of Beta-Alanine Synthase From Saccharomyces Kluyveri
Organism: Saccharomyces kluyveri
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2007-10-02 Classification: HYDROLASE Ligands: ZN, URP, DTT |
 |
Organism: Lachancea kluyveri
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-03-14 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of Dihydropyrimidinase From Saccharomyces Kluyveri In Complex With The Substrate Dihydrouracil
Organism: Lachancea kluyveri
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-03-14 Classification: HYDROLASE Ligands: ZN, DUC |
 |
Crystal Structure Of Dihydropyrimidinase From Saccharomyces Kluyveri In Complex With The Reaction Product N-Carbamyl-Beta-Alanine
Organism: Lachancea kluyveri
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2006-03-14 Classification: HYDROLASE Ligands: ZN, URP |
 |
Organism: Lachancea kluyveri
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2003-11-11 Classification: HYDROLASE Ligands: ZN, BIB |