Search Count: 1,353
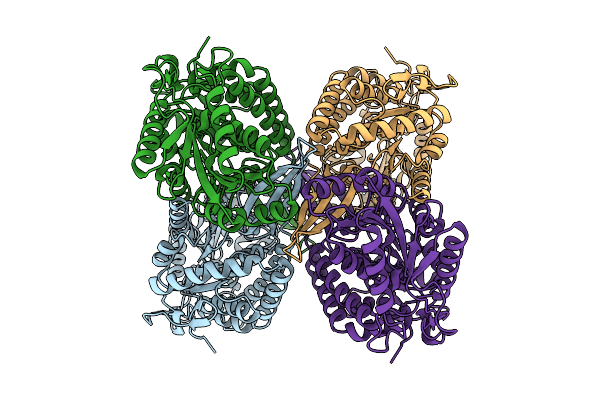 |
Cryoem Structure Of Aldehyde Dehydrogenase From Burkholderia Cenocepacia At 2.33A Resolution
Organism: Burkholderia cenocepacia
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: OXIDOREDUCTASE |
 |
Organism: Burkholderia cenocepacia, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
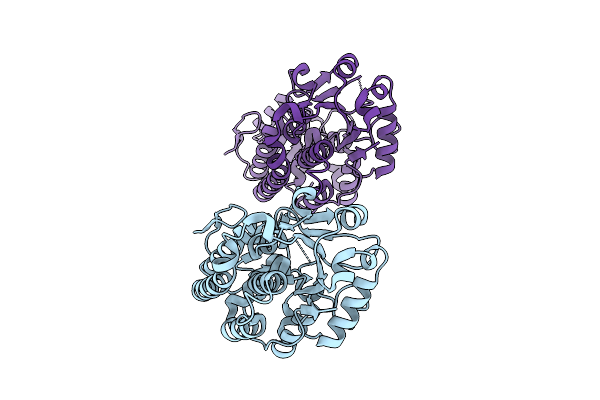 |
Organism: Yersinia ruckeri
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: METAL BINDING PROTEIN |
 |
Structure Of Yiua From Yersinia Ruckeri With Iron And Nitrilotriacetic Acid
Organism: Yersinia ruckeri
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: METAL BINDING PROTEIN Ligands: NTA, FE |
 |
Organism: Yersinia ruckeri
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Organism: Yersinia ruckeri
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: METAL BINDING PROTEIN Ligands: FE, A1I15, SO4 |
 |
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: CELL ADHESION Ligands: ACT, GOL |
 |
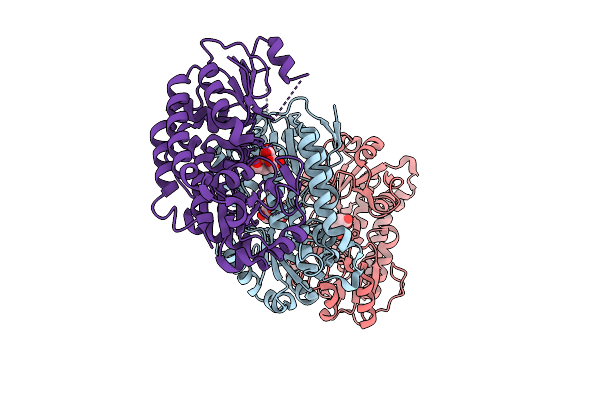
Cryoem Structure Of Methylmalonic Acid Semialdehyde Dehydrogenase From Burkholderia Cenocepacia At 2.38A Resolution
Organism: Burkholderia cenocepacia
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
The Structure Of Natural P450Bm3-H Derived From Bacillus Megaterium For Catalyzing The Steroid Dhea
Organism: Priestia megaterium nbrc 15308 = atcc 14581
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2025-05-07 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Synchrotron X-Ray Crystal Structure Of Oxygen-Bound F87A/F393H P450Bm3 With Decoy C7Prophe (N-Enanthyl-L-Prolyl-L-Phenylalanine) And Substrate Styrene At 2 Mgy X-Ray Dose
Organism: Priestia megaterium nbrc 15308 = atcc 14581
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: HEM, SYN, OXY, D0L, GOL |
 |
Xfel Crystal Structure Of The Oxidized Form Of F393H P450Bm3 With N-Enanthyl-L-Prolyl-L-Phenylalanine In Complex With Styrene
Organism: Priestia megaterium nbrc 15308 = atcc 14581
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: HEM, D0L, SYN, GOL |
 |
Xfel Crystal Structure Of The Oxidized Form Of F87A/F393H P450Bm3 With N-Enanthyl-L-Prolyl-L-Phenylalanine In Complex With Styrene
Organism: Priestia megaterium nbrc 15308 = atcc 14581
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: HEM, D0L, SYN, GOL |
 |
Xfel Crystal Structure Of The Reduced Form Of F393H P450Bm3 With N-Enanthyl-L-Prolyl-L-Phenylalanine In Complex With Styrene
Organism: Priestia megaterium nbrc 15308 = atcc 14581
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: HEM, D0L, SYN, GOL |
 |
Xfel Crystal Structure Of The Reduced Form Of F87A/F393H P450Bm3 With N-Enanthyl-L-Prolyl-L-Phenylalanine In Complex With Styrene
Organism: Priestia megaterium nbrc 15308 = atcc 14581
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: HEM, D0L, SYN, GOL |
 |
Xfel Crystal Structure Of The Oxygen-Bound Form Of F393H P450Bm3 With N-Enanthyl-L-Prolyl-L-Phenylalanine In Complex With Styrene
Organism: Priestia megaterium nbrc 15308 = atcc 14581
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: HEM, D0L, SYN, OXY, GOL |
 |
Xfel Crystal Structure Of The Oxygen-Bound Form Of F87A/F393H P450Bm3 With N-Enanthyl-L-Prolyl-L-Phenylalanine In Complex With Styrene
Organism: Priestia megaterium nbrc 15308 = atcc 14581
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: HEM, D0L, SYN, OXY, GOL |
 |
Cryo-Em Structure Of The Inhibitor-Bound Vo Complex From Enterococcus Hirae
Organism: Enterococcus hirae atcc 9790
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CDL, NA, W3K |
 |
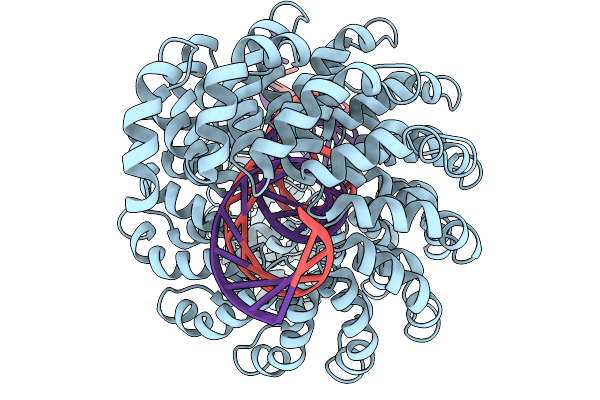
Crystal Structure Of Dna N6-Adenine Methyltransferase M.Bcejiv From Burkholderia Cenocepacia In Complex With Duplex Dna Substrates
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-09-11 Classification: DNA BINDING PROTEIN Ligands: EPE, SFG |
 |
Crystal Structure Of Dna N6-Adenine Methyltransferase M.Bcejiv From Burkholderia Cenocepacia In Complex With Duplex Dna Substrate Containing Gtatac As Recognition Sequence
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-09-11 Classification: DNA BINDING PROTEIN Ligands: SFG |
 |
Crystal Structure Of Dna N6-Adenine Methyltransferase M.Bcejiv From Burkholderia Cenocepacia In Complex With Duplex Dna Substrate Containing Gtaaac As Recognition Sequence
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2024-09-11 Classification: DNA BINDING PROTEIN Ligands: SFG |

