Search Count: 33,290
 |
Organism: Faecalibacterium duncaniae
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: TRS, GOL, EDO, PEG |
 |
Organism: Faecalibacterium duncaniae
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: TRS, EDO |
 |
Organism: Schizosaccharomyces pombe 972h-
Method: SOLUTION NMR Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN |
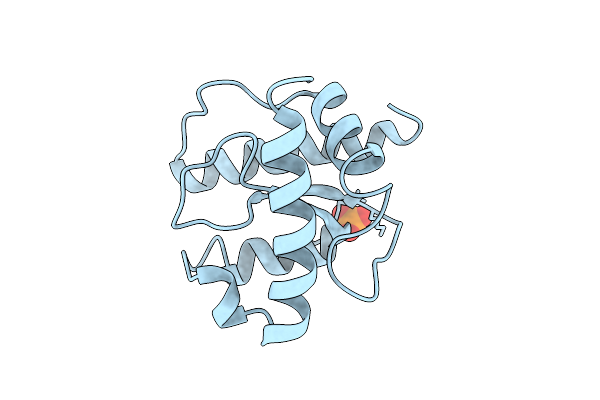 |
X-Ray Structure Of The C-Terminal Domain (Residues 366-485) Of S. Pombe Threonylcarbamoyladenosine Dehydratase
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN Ligands: PO4 |
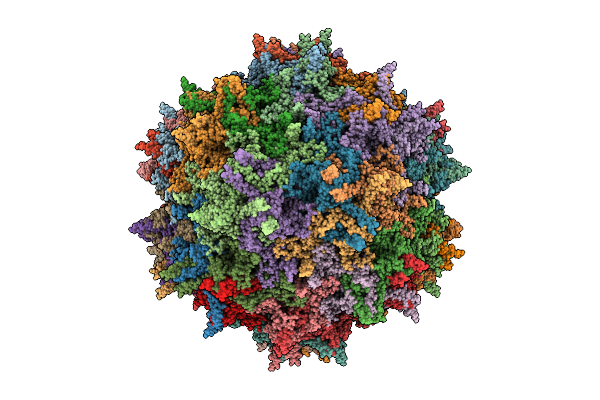 |
Organism: Adeno-associated virus 2 srivastava/1982
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRUS |
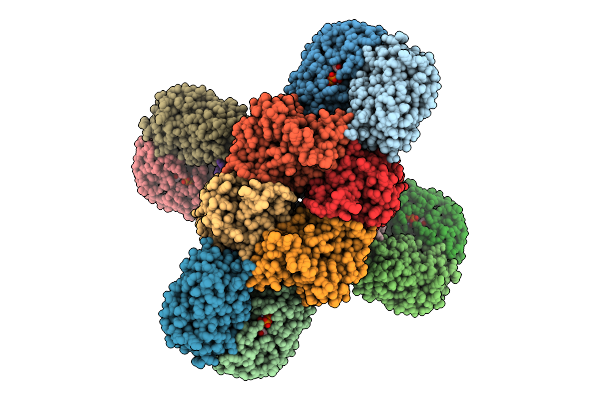 |
Organism: Archangium gephyra
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: CELL INVASION Ligands: Y43, ZN, NAD |
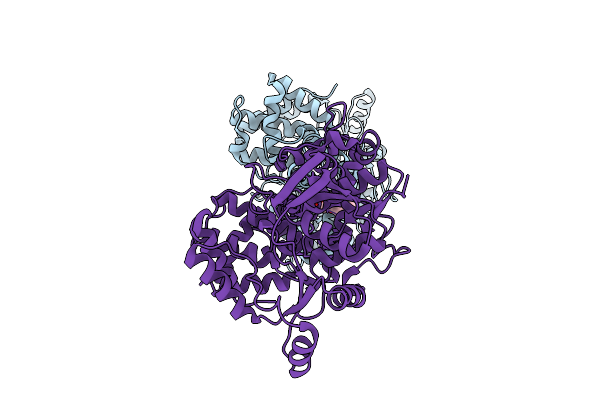 |
Crystal Structure Of Glutamate--Trna Ligase (Gltx) From Moraxella Catarrhalis (Apo)
Organism: Moraxella catarrhalis
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LIGASE Ligands: SO4, P6G, CL |
 |
Organism: Archangium gephyra
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: CELL INVASION |
 |
Organism: Archangium gephyra
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: CELL INVASION Ligands: Y43, ZN, NCA |
 |
Organism: Archangium gephyra
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: CELL INVASION |
 |
Organism: Archangium gephyra
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: CELL INVASION |
 |
Organism: Archangium gephyra
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: CELL INVASION |
 |
Organism: Archangium gephyra
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: CELL INVASION Ligands: Y43, ZN |
 |
Organism: Mesorhizobium metallidurans
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: P6G, PEG, EPE, NA, TRS |
 |
Cryoem Structure Of Aldehyde Dehydrogenase From Burkholderia Cenocepacia At 2.33A Resolution
Organism: Burkholderia cenocepacia
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: OXIDOREDUCTASE |
 |
Organism: Leptospirillum ferriphilum
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: DNA BINDING PROTEIN |
 |
Organism: Candida parapsilosis
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: LYASE Ligands: ZN, AZM |
 |
Organism: Burkholderia cenocepacia, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
Organism: Streptomyces roseifaciens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: BIOSYNTHETIC PROTEIN Ligands: VVO, SIN |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: DNA BINDING PROTEIN Ligands: ACY, SO4, HCI |

