Search Count: 24
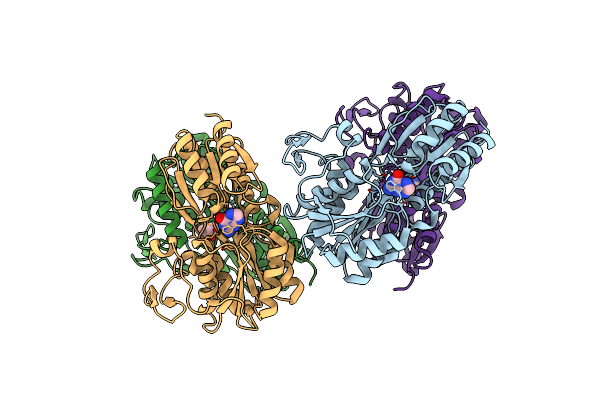 |
Organism: Pseudomonas sp. (strain adp)
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2019-11-13 Classification: TRANSPORT PROTEIN Ligands: GUN, DMS |
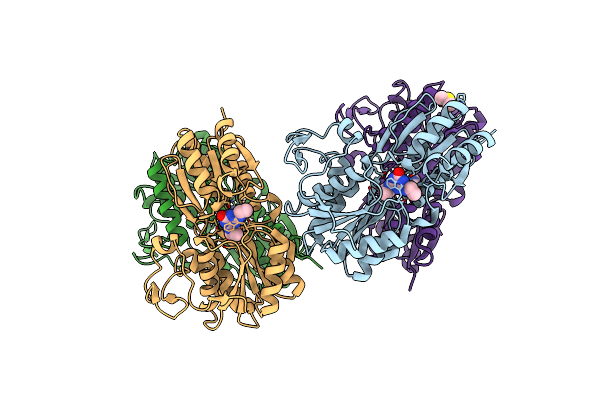 |
Organism: Pseudomonas sp. (strain adp)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-11-13 Classification: TRANSPORT PROTEIN Ligands: OKM, DMS |
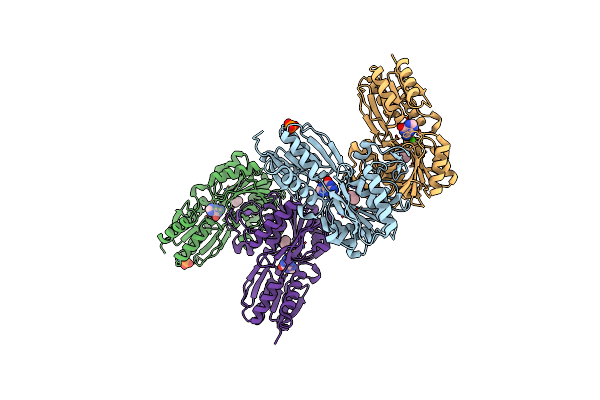 |
Organism: Pseudomonas sp. (strain adp)
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2019-11-13 Classification: TRANSPORT PROTEIN Ligands: GUN, EMC, PO4, CA |
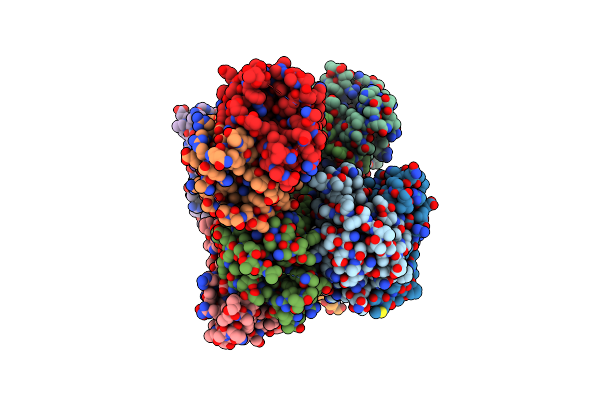 |
The Structure Of Atzh: A Little Known Member Of The Atrazine Breakdown Pathway
Organism: Pseudomonas sp. (strain adp)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-11-14 Classification: UNKNOWN FUNCTION Ligands: 6JN |
 |
X-Ray Structure And Mutagenesis Studies Of The N-Isopropylammelide Isopropylaminohydrolase, Atzc
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-03-18 Classification: HYDROLASE Ligands: CL, ZN |
 |
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-03-11 Classification: HYDROLASE Ligands: FE, PEG |
 |
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-03-11 Classification: HYDROLASE Ligands: FE, EDO, PEG, CL |
 |
The Reaction Mechanism Of The N-Isopropylammelide Isopropylaminohydrolase Atzc: Insights From Structural And Mutagenesis Studies
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: ZN, CL, DMS, MLI |
 |
The Reaction Mechanism Of The N-Isopropylammelide Isopropylaminohydrolase Atzc: Insights From Structural And Mutagenesis Studies
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: ZN, CL, EPE |
 |
The Reaction Mechanism Of The N-Isopropylammelide Isopropylaminohydrolase Atzc: Insights From Structural And Mutagenesis Studies
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2015-03-04 Classification: HYDROLASE Ligands: ZN, MES, CL, MLI |
 |
Structure Of The Amidase Domain Of Allophanate Hydrolase From Pseudomonas Sp Strain Adp
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-11-26 Classification: HYDROLASE Ligands: MLI |
 |
Cyanuric Acid Hydrolase: Evolutionary Innovation By Structural Concatenation.
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-07-17 Classification: HYDROLASE Ligands: PO4, MG, PEG |
 |
Cyanuric Acid Hydrolase: Evolutionary Innovation By Structural Concatenation.
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2013-07-17 Classification: HYDROLASE Ligands: MG, WDL, PEG |
 |
Cyanuric Acid Hydrolase: Evolutionary Innovation By Structural Concatenation.
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-07-17 Classification: HYDROLASE Ligands: MG, AX2 |
 |
Cyanuric Acid Hydrolase: Evolutionary Innovation By Structural Concatenation
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-07-17 Classification: HYDROLASE Ligands: MG, BR8, PEG |
 |
Crystal Structure Of N-Isopropylammelide Isopropylaminohydrolase Atzc From Pseudomonas Sp. Strain Adp Complexed With Zn
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2007-09-11 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Phyllocnistis sp. BOLD:AAI3014
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Pseudomonas sp. (strain ADP)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Pseudomonas sp. (strain ADP)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
Organism: Pseudomonas sp. (strain ADP)
Method: Alphafold Release Date: Classification: NA Ligands: NA |

