Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
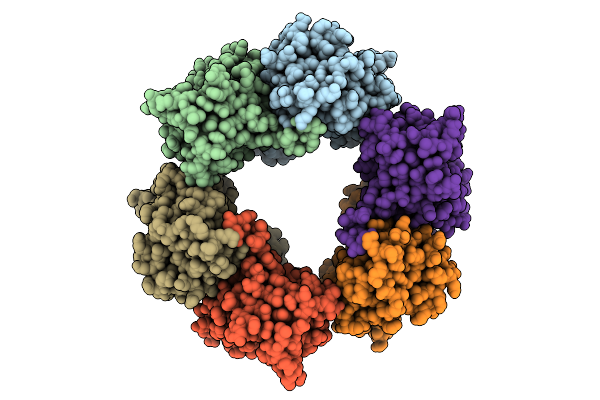 |
Cryo-Em Structure Of Candida Albicans Ph Regulated Antigen 1 (Pra1) Protein In The Absence Of Zn2+
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: METAL BINDING PROTEIN |
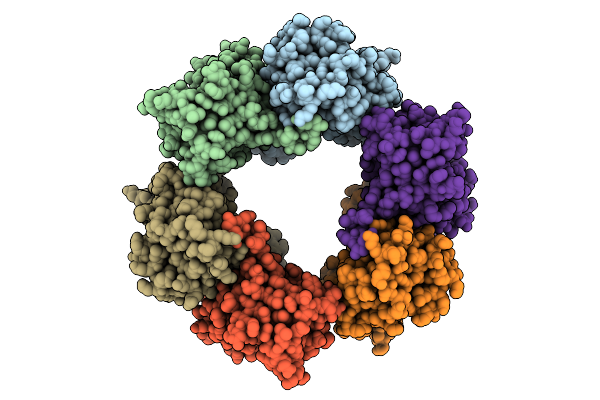 |
Cryo-Em Structure Of Candida Albicans Ph Regulated Antigen 1 (Pra1) Protein In Complex With Zn2+
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: METAL BINDING PROTEIN Ligands: ZN |
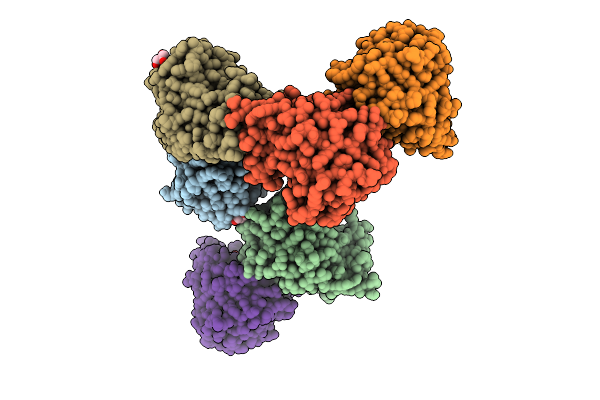 |
Crystal Structure Of Bacteroides Ovatus Kdui1 Responsible For Metabolism Of Glycosaminoglycan
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: ISOMERASE Ligands: GOL |
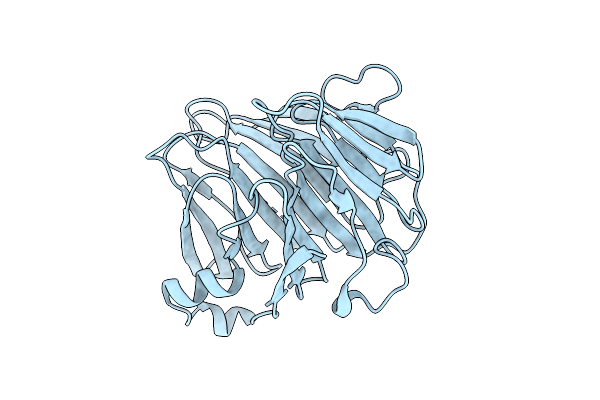 |
Crystal Structure Of Bacteroides Ovatus Kdui2 Responsible For Metabolism Of Glycosaminoglycan
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: ISOMERASE |
 |
Crystal Structure Of Bacteroides Ovatus Dhud Responsible For Metabolism Of Glycosaminoglycan
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: OXIDOREDUCTASE Ligands: ACT |
 |
Crystal Structure Of Bacteroides Ovatus Dhud Complexed With Nad+ Responsible For Metabolism Of Glycosaminoglycan
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSFERASE/INHIBITOR Ligands: PY1, CL |
 |
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSFERASE/INHIBITOR Ligands: CL, A1B7R, SO4 |
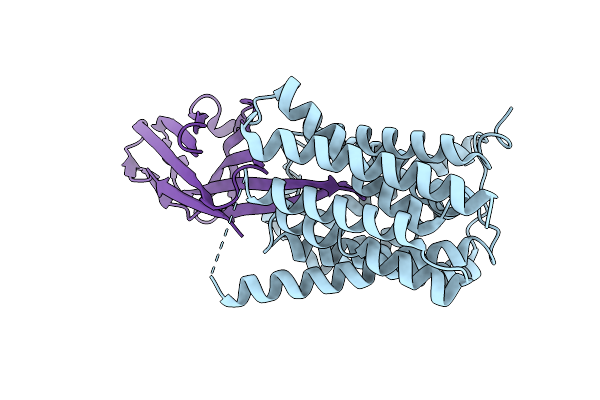 |
Cryo-Em Structure Of Candida Albicans Vrg4 Bound To An Inhibitory Nanobody.
Organism: Lama glama, Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN |
 |
Cryo-Em Structure Of Candida Albicans Vrg4 Bound To An Inhibitory Nanobody And Gdp-Mannose.
Organism: Candida albicans, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN Ligands: GDD |
 |
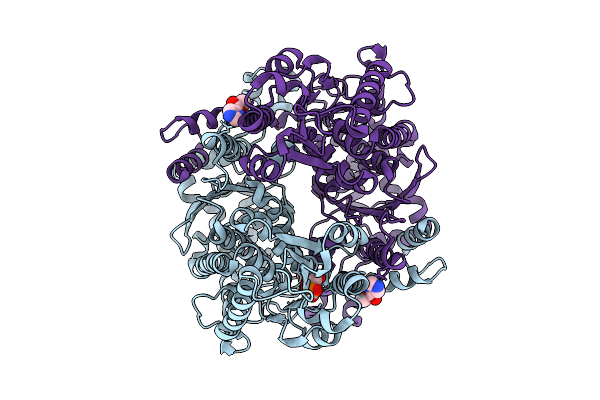
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With Fragments
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ISOMERASE Ligands: PA5, A1IHU |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With Fragments
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ISOMERASE Ligands: A1IIB, PA5, CL |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With A Fragment
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: ISOMERASE Ligands: PA5, A1IH9, CL |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerae (Capgi) In Complex With Fragments
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: ISOMERASE Ligands: PA5, A1IIA, A1IH8, CL |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With A Fragment Binder
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: ISOMERASE Ligands: PA5, A1IIC |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With Fructose-6-Phosphate
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: ISOMERASE Ligands: F6P, CL |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With A Fragment
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: ISOMERASE Ligands: A1IHU, PA5 |
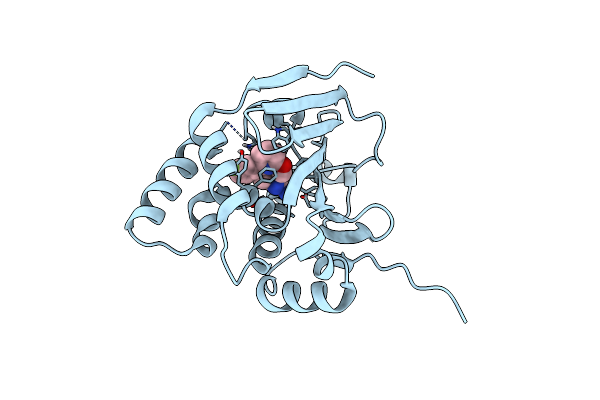 |
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: CHAPERONE/INHIBITOR Ligands: 990 |
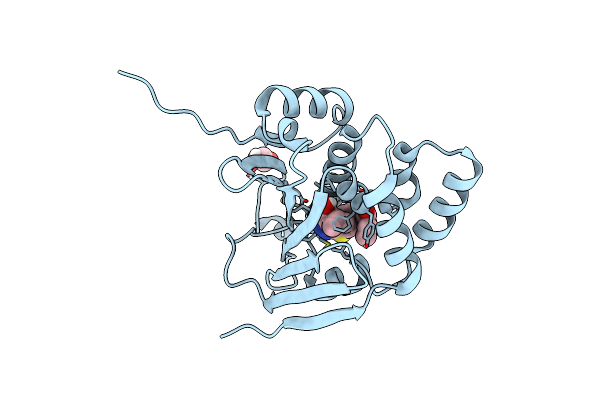 |
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: CHAPERONE/INHIBITOR Ligands: 819, EDO |
 |
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: CHAPERONE/INHIBITOR Ligands: 2KL |