Search Count: 1,209
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: PLANT PROTEIN Ligands: FAD |
 |
Organism: Zea mays
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: CHOLINE-BINDING PROTEIN Ligands: URE |
 |
Organism: Zea mays
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PLANT PROTEIN Ligands: COA |
 |
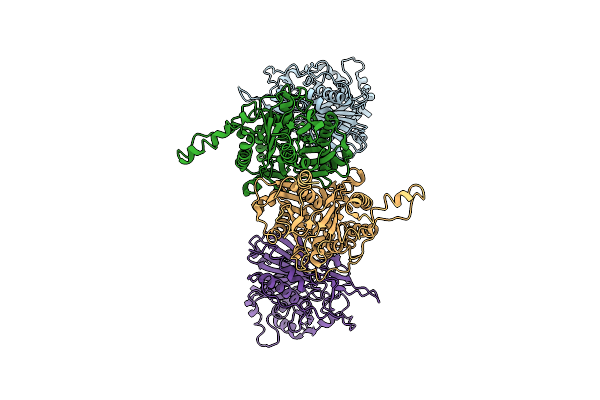
Cryo-Em Structure Of The Maize Cer6-Gl2 Complex (Inactive C222A Mutant) Bound With Malonyl-Coa
Organism: Zea mays
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PLANT PROTEIN Ligands: MLC |
 |
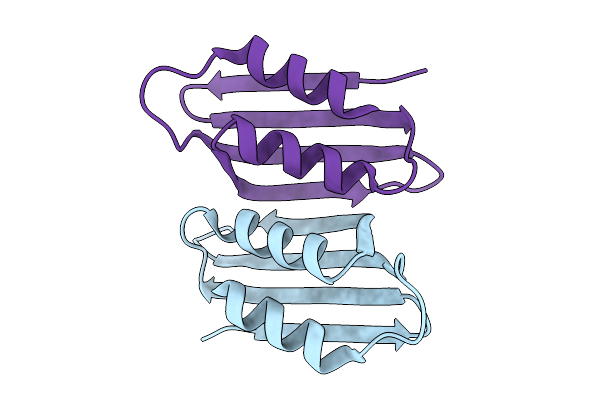
Cryo-Em Structure Of The Maize Cer6-Gl2 Complex In The Presence Of 30:0 Coa
Organism: Zea mays
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PLANT PROTEIN |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: LIPID BINDING PROTEIN |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: OXIDOREDUCTASE |
 |
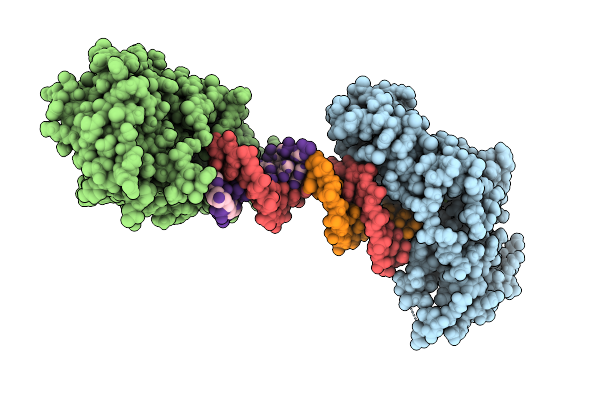
Crystal Structure Of Zea Mays 3-Phosphoglycerate Dehydrogenase S282L Mutant
Organism: Zea mays
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: OXIDOREDUCTASE |
 |
Organism: Zea mays, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN/DNA Ligands: MG |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PLANT PROTEIN Ligands: MN |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: NAP, EDO |
 |
Crystal Structure Of Zmet2 In Complex With Unmethylated Ctg Dna And A Histone H3Kc9Me2 Peptide
Organism: Zea mays, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2025-05-21 Classification: TRANSFERASE Ligands: SAH |
 |
Crystal Structure Of Zea Mays Adenosine Kinase 3 (Zmadk3) In Complex With Ap5A
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2025-01-29 Classification: TRANSFERASE Ligands: AP5, EDO, PEG, CL |
 |
Neutron Structure Of Ferredoxin-Nadp+ Reductase From Maize Root -Reduced Form
Organism: Zea mays
Method: NEUTRON DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: MES, FDA |
 |
Neutron Structure Of Ferredoxin-Nadp+ Reductase From Maize Root -Oxidized Form
Organism: Zea mays
Method: NEUTRON DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: FAD, MES |
 |
High Resolution Structure Of Ferredoxin-Nadp+ Reductase From Maize Root - Oxidized Form, Low X-Ray Dose
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: MES, GOL, FAD |
 |
High Resolution Structure Of Ferredoxin-Nadp+ Reductase From Maize Root - Reduced Form, Low X-Ray Dose
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: MES, FDA |
 |
R115A Mutant Of Ferredoxin-Nadp+ Reductase From Maize Root - Oxidized Form, Low X-Ray Dose
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: FAD, MES |
 |
Organism: Kutzneria buriramensis
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2025-01-15 Classification: HYDROLASE |
 |
Organism: Kutzneria buriramensis
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2025-01-15 Classification: HYDROLASE |

