Search Count: 1,847
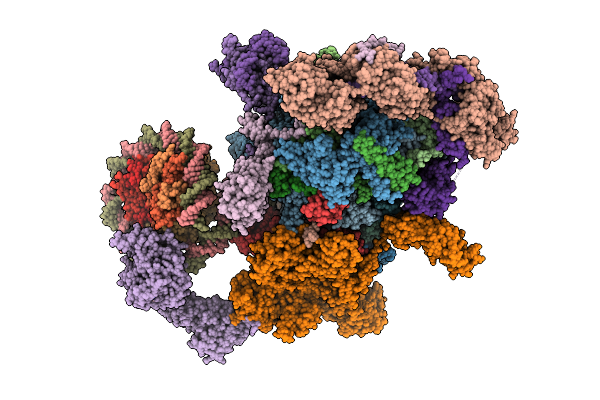 |
Co-Transcriptional Histone H3K36 Methylation Complex Containing Rna Polymerase Ii Elongation Complex, Set2, And The Upstream Nucleosome. (Temp115, Type B)
Organism: Komagataella phaffii gs115, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSCRIPTION Ligands: ZN, MG, SAH |
 |
Co-Transcriptional Histone H3K36 Methylation Complex Containing Rna Polymerase Ii Elongation Complex, Set2, And The Upstream Nucleosome. (Temp115, Type A)
Organism: Komagataella phaffii gs115, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSCRIPTION Ligands: ZN, MG, SAH |
 |
Co-Transcriptional Histone H3K36 Methylation Complex Containing Rna Polymerase Ii Elongation Complex, Set2, And The Upstream Nucleosome. (Temp130, Type A)
Organism: Komagataella phaffii (strain gs115 / atcc 20864), Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSCRIPTION Ligands: ZN, MG, SAH |
 |
Co-Transcriptional Histone H3K36 Methylation Complex Containing Rna Polymerase Ii Elongation Complex, Set2, And The Upstream Nucleosome. (Temp130, Type B)
Organism: Komagataella phaffii gs115, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSCRIPTION Ligands: ZN, MG, SAH |
 |
Co-Transcriptional Histone H3K36 Methylation Complex Containing Rna Polymerase Ii Elongation Complex, Set2, And The Upstream Nucleosome. (Temp115, Fact-Hexamer)
Organism: Komagataella phaffii gs115, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSCRIPTION Ligands: ZN, MG |
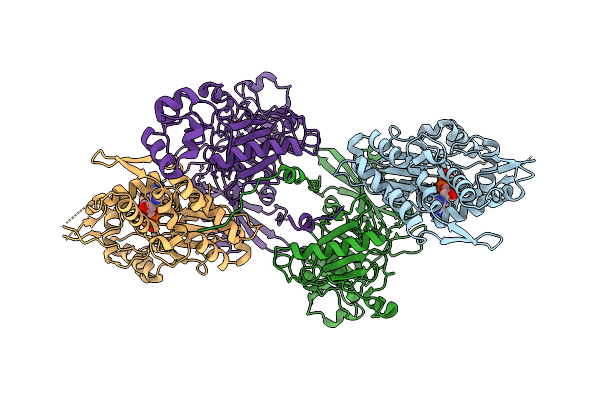 |
Organism: Paralvinella sulfincola, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:3.29 Å Release Date: 2025-10-15 Classification: STRUCTURAL PROTEIN Ligands: CA, ADP |
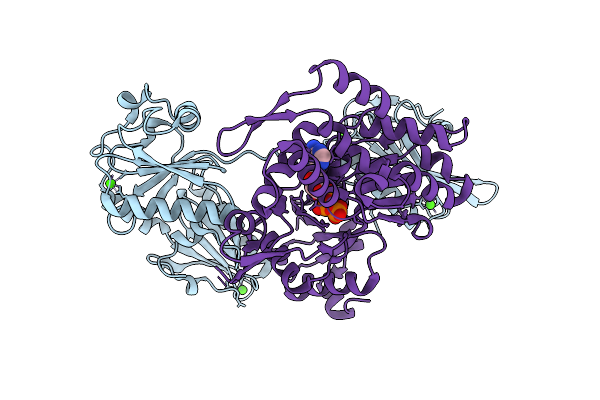 |
Organism: Paralvinella sulfincola, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2025-10-15 Classification: STRUCTURAL PROTEIN Ligands: CA, ATP |
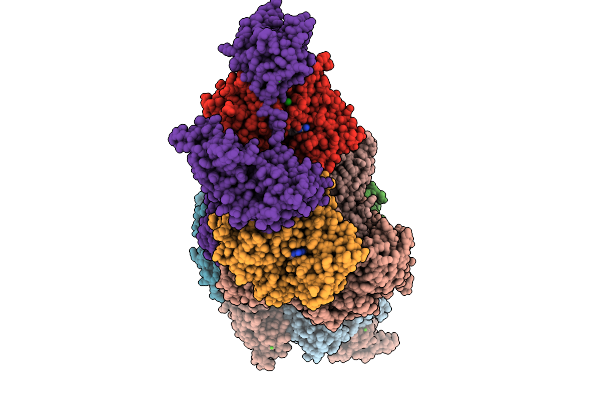 |
Organism: Gallus gallus, Paralvinella sulfincola
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-10-08 Classification: STRUCTURAL PROTEIN Ligands: ADP, MG, CA, CL, SCN |
 |
Organism: Gluconobacter oxydans
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: EDO, CIT |
 |
The X-Ray Crystallographic Structure Of Oligo-1,6-Glucosidase From Paenibacillus Sp. Stb16
Organism: Paenibacillus
Method: X-RAY DIFFRACTION Resolution:3.24 Å Release Date: 2025-05-07 Classification: HYDROLASE |
 |
Structure Of Bacillus Phage Spo1 Anti-Cbass 4 (Acb4) In Complex With 3'3'-Cgamp
Organism: Bacillus phage spo1
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2025-01-29 Classification: VIRAL PROTEIN Ligands: 4BW |
 |
Organism: Leptospira interrogans serovar copenhageni
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-12-18 Classification: SIGNALING PROTEIN Ligands: ACP, MG, BEF, SO4 |
 |
Organism: Bacillus, Bacillus phage spo1
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: TRANSCRIPTION |
 |
Organism: Bacillus, Bacillus phage spo1
Method: ELECTRON MICROSCOPY Resolution:2.94 Å Release Date: 2024-12-04 Classification: TRANSCRIPTION |
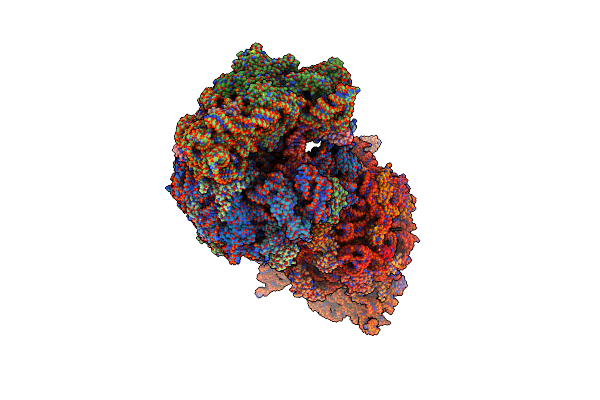 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Lasso Peptide Lariocidin, Mrna, Aminoacylated A-Site Phe-Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.50A Resolution
Organism: Escherichia coli, Paenibacillus, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, K, ZN, SF4 |
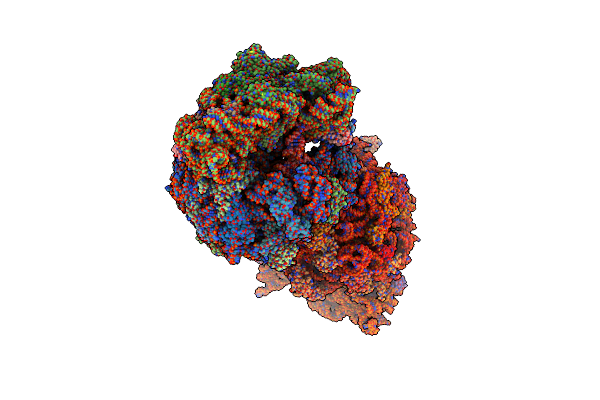 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Lasso Peptide Lariocidin B, Mrna, Aminoacylated A-Site Phe-Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.60A Resolution
Organism: Escherichia coli, Paenibacillus, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, K, ZN, SF4 |
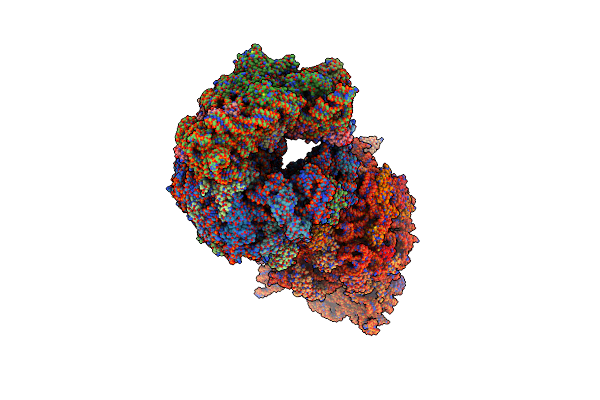 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Lasso Peptide Lariocidin And Protein Y At 2.60A Resolution
Organism: Escherichia coli k-12, Paenibacillus, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, ARG, MPD, ZN, SF4 |
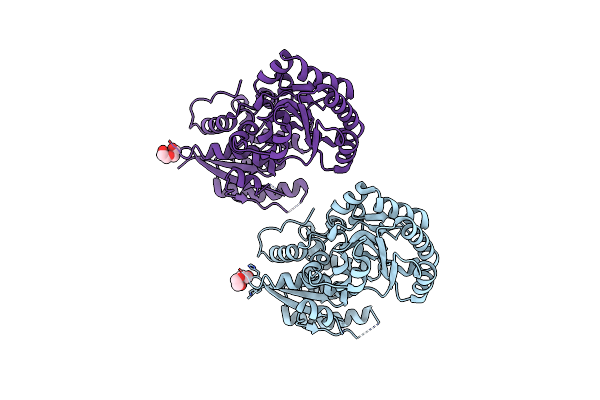 |
Organism: Paenibacillus alvei
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-10-02 Classification: ISOMERASE Ligands: PEG |
 |
Organism: Bacillus phage spo1
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2024-09-25 Classification: VIRAL PROTEIN |
 |
Phage Spo1 Protein Gp49 Is A Novel Rna Binding Protein That Involves In Host Iron Metabolism
Organism: Bacillus phage spo1
Method: SOLUTION NMR Release Date: 2024-09-04 Classification: RNA BINDING PROTEIN |

