Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) T40C-S315C (Db1) Variant In Complex With Prenylated Flavin Hydroxylated At The C1 Prime Position
Organism: Aspergillus niger cbs 513.88
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2023-09-06 Classification: LYASE Ligands: BYN, MN, K, SCN |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) S145C-P289C (Db2) Variant In Complex With Prenylated Flavin Hydroxylated At The C1 Prime Position
Organism: Aspergillus niger cbs 513.88
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2023-09-06 Classification: LYASE Ligands: BYN, MN, K, SCN |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) C122-S261C (Db3) Variant In Complex With Prenylated Flavin
Organism: Aspergillus niger cbs 513.88
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2023-09-06 Classification: LYASE Ligands: 4LU, MN, K, SCN |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) V186C-A296C (Db4) Variant In Complex With Prenylated Flavin
Organism: Aspergillus niger cbs 513.88
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-09-06 Classification: LYASE Ligands: 4LU, MN, K, SCN |
 |
Organism: Aspergillus niger cbs 513.88
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2021-09-22 Classification: HYDROLASE Ligands: ZN, MLI, CL |
 |
Organism: Aspergillus niger (strain cbs 513.88 / fgsc a1513)
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2021-08-04 Classification: LYASE Ligands: K, 4LU, MN |
 |
Structure Of A. Niger Fdc T395M R435P P438W Variant (Anfdcii) In Complex With Prfmn
Organism: Aspergillus niger (strain cbs 513.88 / fgsc a1513)
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2021-08-04 Classification: LYASE Ligands: MN, K, BYN |
 |
Structure Of Wild Type A. Niger Fdc1 Purified In The Dark With Prfmn In The Iminium Form
Organism: Aspergillus niger (strain cbs 513.88 / fgsc a1513)
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2017-12-27 Classification: LYASE Ligands: 4LU, MN, K |
 |
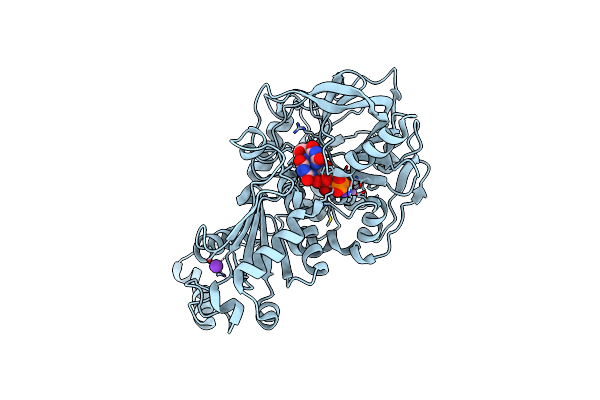
Crystal Structure Of E282Q A. Niger Fdc1 With Prfmn In The Hydroxylated Form
Organism: Aspergillus niger (strain cbs 513.88 / fgsc a1513)
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2017-12-20 Classification: LYASE Ligands: MN, K, BYN, SCN |
 |
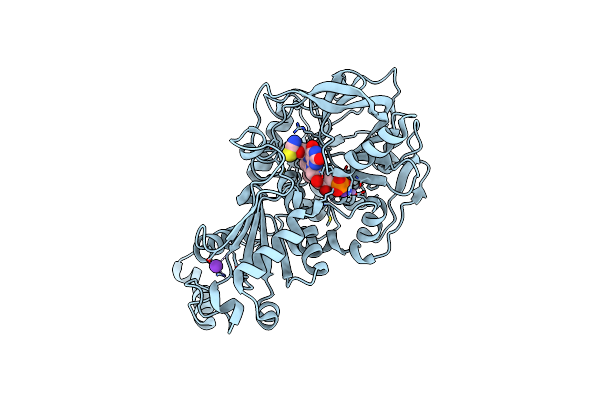
Structure Of E282Q A. Niger Fdc1 With Prfmn In The Hydroxylated And Ketimine Forms
Organism: Aspergillus niger (strain cbs 513.88 / fgsc a1513)
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2017-12-20 Classification: LYASE Ligands: MN, K, FZZ, BYN |
 |
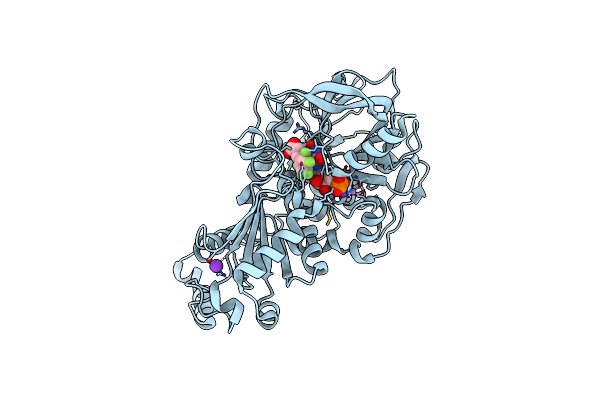
Organism: Aspergillus niger (strain cbs 513.88 / fgsc a1513)
Method: X-RAY DIFFRACTION Resolution:1.03 Å Release Date: 2017-12-20 Classification: LYASE Ligands: MN, K, BYN, SCN |
 |
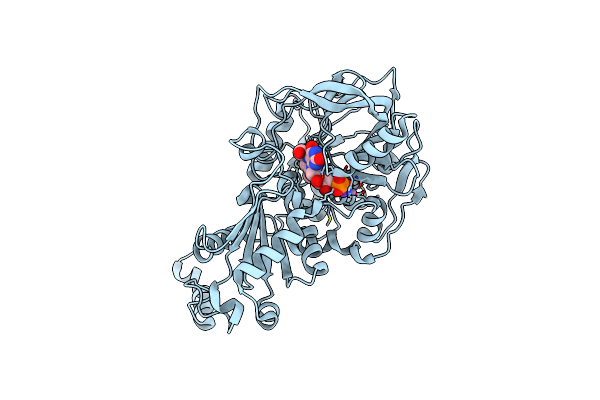
Organism: Aspergillus niger (strain cbs 513.88 / fgsc a1513)
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2017-12-20 Classification: LYASE Ligands: 4LU, MN, K, F5C |
 |
Organism: Aspergillus niger (strain cbs 513.88 / fgsc a1513)
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2017-12-20 Classification: LYASE Ligands: MN, K, BYN |
 |
Organism: Aspergillus niger cbs 513.88
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2017-04-19 Classification: HYDROLASE Ligands: NAG, SO4, CL, BTB, GOL |
 |
Structure Of E298Q-Beta-Galactosidase From Aspergillus Niger In Complex With 3-B-Galactopyranosyl Glucose
Organism: Aspergillus niger cbs 513.88
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2017-04-19 Classification: HYDROLASE Ligands: NAG, SO4, CL, DMS |
 |
Structure Of E298Q-Beta-Galactosidase From Aspergillus Niger In Complex With Allolactose
Organism: Aspergillus niger cbs 513.88
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-04-19 Classification: HYDROLASE Ligands: NAG, SO4, CL, DMS |
 |
Structure Of E298Q-Beta-Galactosidase From Aspergillus Niger In Complex With 6-B-Galactopyranosyl Galactose
Organism: Aspergillus niger cbs 513.88
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2017-04-19 Classification: HYDROLASE Ligands: NAG, CL, 1PE |
 |
Structure Of E298Q-Beta-Galactosidase From Aspergillus Niger In Complex With 4-Galactosyl-Lactose
Organism: Aspergillus niger cbs 513.88
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-04-19 Classification: HYDROLASE Ligands: NAG |
 |
Structure Of E298Q-Beta-Galactosidase From Aspergillus Niger In Complex With 6-Galactosyl-Lactose
Organism: Aspergillus niger cbs 513.88
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-04-19 Classification: HYDROLASE Ligands: NAG, DMS, CL, 1PE |
 |
Structure Of A. Niger Fdc1 With The Prenylated-Flavin Cofactor In The Iminium Form.
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2015-06-17 Classification: LYASE Ligands: 4LU, MN, K |