Search Count: 207
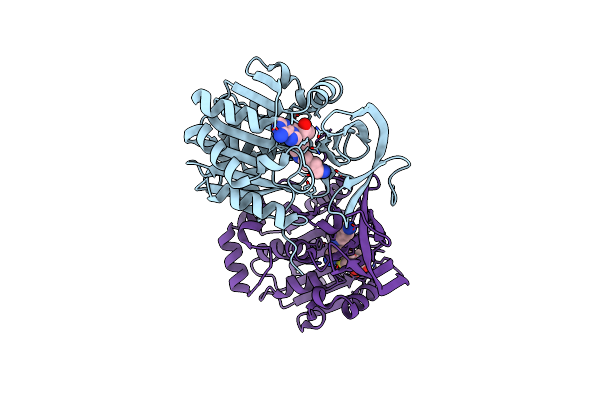 |
Crystal Structure Of The Aminopropyltransferase, Spee From Hyperthermophilic Crenarchaeon, Pyrobaculum Calidifontis In Complex With 5'-Methylthioadenosine (Mta) And Spermidine
Organism: Pyrobaculum calidifontis jcm 11548
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-06-15 Classification: TRANSFERASE Ligands: MTA, SPD |
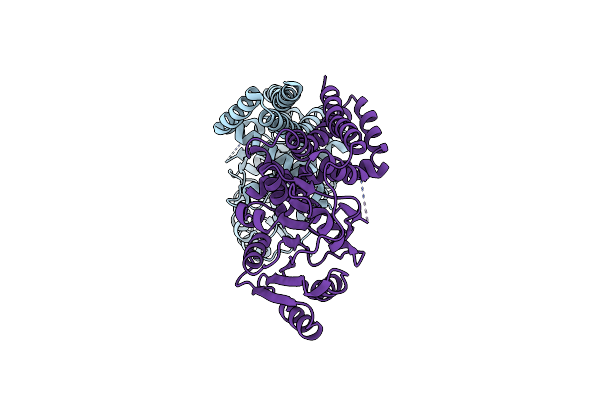 |
Organism: Pyrobaculum calidifontis (strain jcm 11548 / va1)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-07-22 Classification: TRANSFERASE |
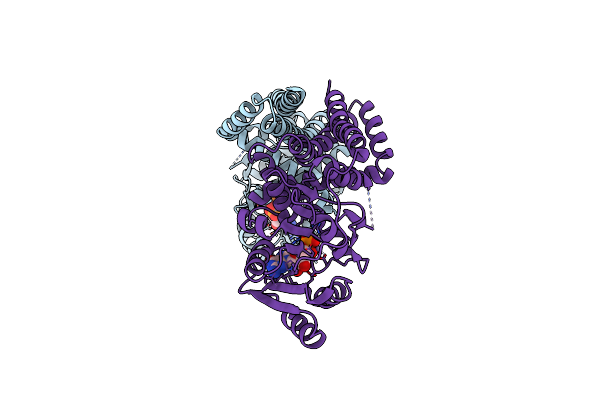 |
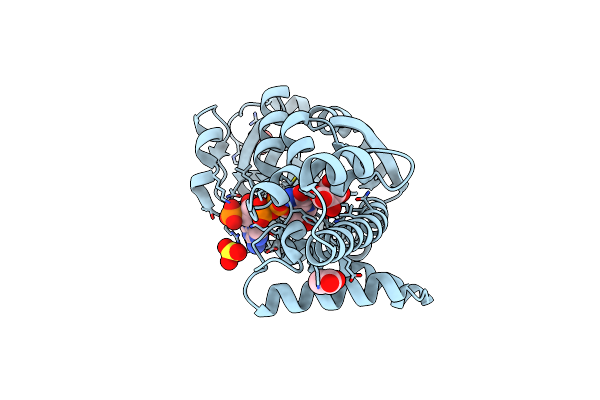
Mannosyltransferase Pcmangt From Pyrobaculum Calidifontis In Complex With Gdp And Mn2+
Organism: Pyrobaculum calidifontis (strain jcm 11548 / va1)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-07-22 Classification: TRANSFERASE Ligands: GDP, MN |
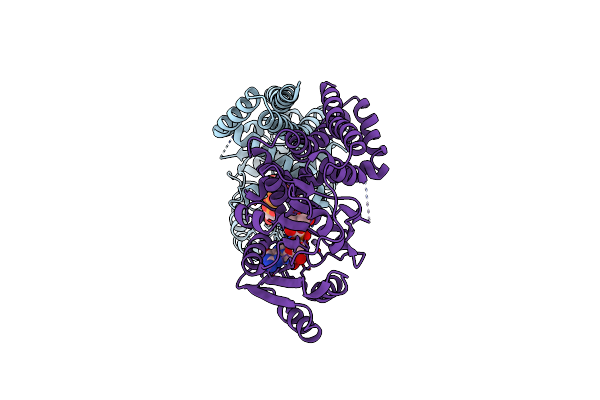 |
Mannosyltransferase Pcmangt From Pyrobaculum Calidifontis In Complex With Gdp-Man And Mn2+
Organism: Pyrobaculum calidifontis (strain jcm 11548 / va1)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-07-22 Classification: TRANSFERASE Ligands: GDD, MN |
 |
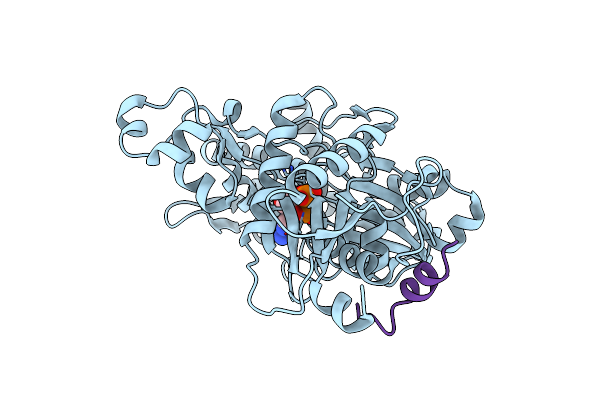
Crystal Structure Of The Nadp+ And Tartrate-Bound Complex Of L-Serine 3-Dehydrogenase From The Hyperthermophilic Archaeon Pyrobaculum Calidifontis
Organism: Pyrobaculum calidifontis (strain jcm 11548 / va1)
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2018-02-07 Classification: OXIDOREDUCTASE Ligands: TLA, NAP, ACY, SO4 |
 |
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-11-30 Classification: STRUCTURAL PROTEIN Ligands: ADP |
 |
Crystal Structure Of The Covalent Thioimide Intermediate Of The Archaeosine Synthase Quef-Like
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2016-11-09 Classification: TRANSFERASE Ligands: GD1, NA |
 |
Organism: Pyrobaculum calidifontis (strain jcm 11548 / va1)
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2016-11-09 Classification: TRANSFERASE Ligands: SCN, PGE, PG4 |
 |
Organism: Pyrobaculum calidifontis jcm 11548
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2016-10-12 Classification: OXIDOREDUCTASE Ligands: ZN, SO4, PPV, NDP |
 |
Organism: Pyrobaculum calidifontis (strain jcm 11548 / va1)
Method: X-RAY DIFFRACTION Resolution:3.47 Å Release Date: 2016-10-12 Classification: LYASE Ligands: ZN |
 |
The 1.18 A Resolution Structure Of L-Serine 3-Dehydrogenase Complexed With Nadp+ And Sulfate Ion From The Hyperthermophilic Archaeon Pyrobaculum Calidifontis
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2015-03-04 Classification: OXIDOREDUCTASE Ligands: NAP, SO4, ACY |
 |
Crystal Structure And Nuclease Activity Of The Crispr-Associated Cas4 Protein Pcal_0546 From Pyrobaculum Calidifontis Containing A [2Fe-2S] Cluster
Organism: Pyrobaculum calidifontis jcm 11548
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2014-09-17 Classification: HYDROLASE Ligands: MG, FES |
 |
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2014-01-22 Classification: STRUCTURAL PROTEIN Ligands: ADP |
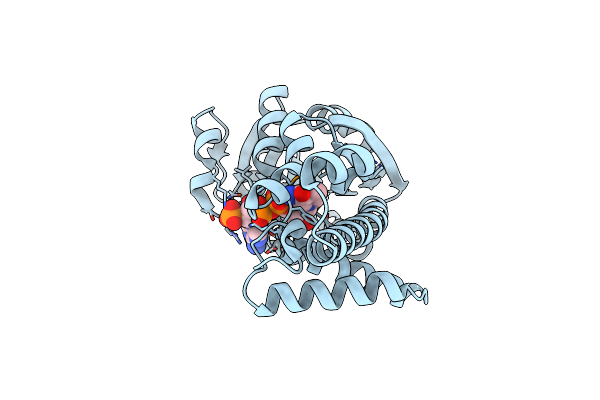 |
Crystal Structure Of Nadp Bound L-Serine 3-Dehydrogenase From Hyperthermophilic Archaeon Pyrobaculum Calidifontis
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-01-15 Classification: OXIDOREDUCTASE Ligands: NAP |
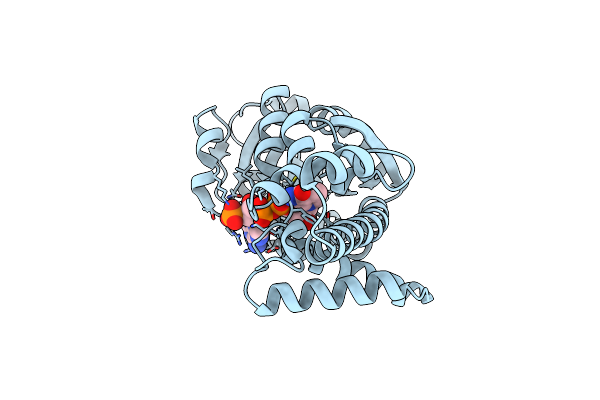 |
Crystal Structure Of Nadp Bound L-Serine 3-Dehydrogenase (K170M) From Hyperthermophilic Archaeon Pyrobaculum Calidifontis
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2014-01-15 Classification: OXIDOREDUCTASE Ligands: NAP |
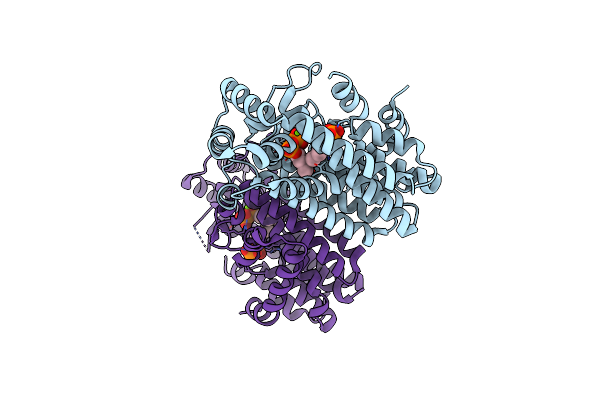 |
Crystal Structure Of Isoprenoid Synthase A3Msh1 (Target Efi-501992) From Pyrobaculum Calidifontis Complexed With Dmapp And Magnesium
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-10-10 Classification: TRANSFERASE Ligands: MG, DMA |
 |
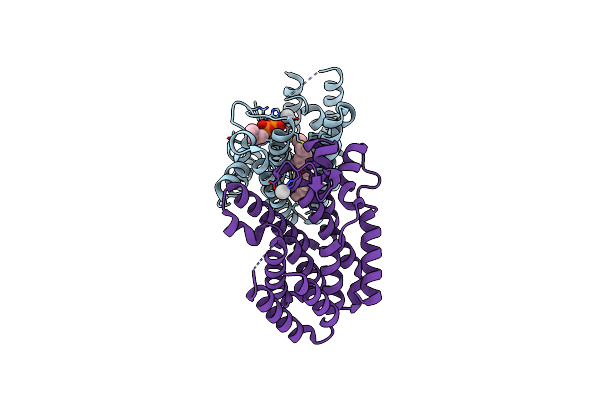
Crystal Structure Of Isoprenoid Synthase A3Msh1 (Target Efi-501992) From Pyrobaculum Calidifontis Complexed With Dmapp
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2012-08-29 Classification: TRANSFERASE Ligands: CL, DMA, PG4 |
 |
Crystal Structure Of Isoprenoid Synthase A3Mx09 (Target Efi-501993) From Pyrobaculum Calidifontis
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-07-11 Classification: TRANSFERASE Ligands: GER, UNX, UNL |
 |
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-02-29 Classification: ISOMERASE Ligands: GDU, NAD |
 |
Crystal Structure Of Isoprenoid Synthase A3Msh1 (Target Efi-501992) From Pyrobaculum Calidifontis
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2012-02-08 Classification: TRANSFERASE Ligands: PO4, CL, ACT |

