Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
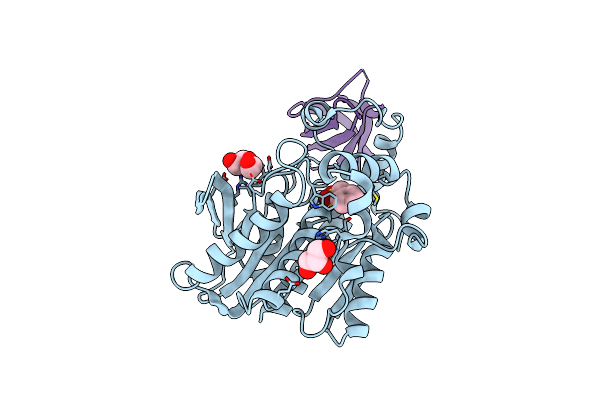 |
Crystal Structure Of The Thermotoga Maritima Cold Shock Protein Mutant Est#13 In Complex With Aacest
Organism: Alicyclobacillus acidocaldarius, Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-15 Classification: PROTEIN BINDING Ligands: PMS, PEG, GOL |
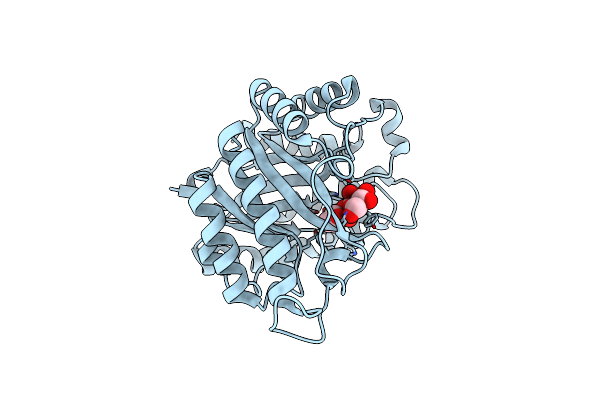 |
The Structure Of Gh113 Beta-Mannanase Aamana From Alicyclobacillus Acidocaldarius In Complex With Manifg
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2014-04-02 Classification: HYDROLASE Ligands: IFM, BMA |
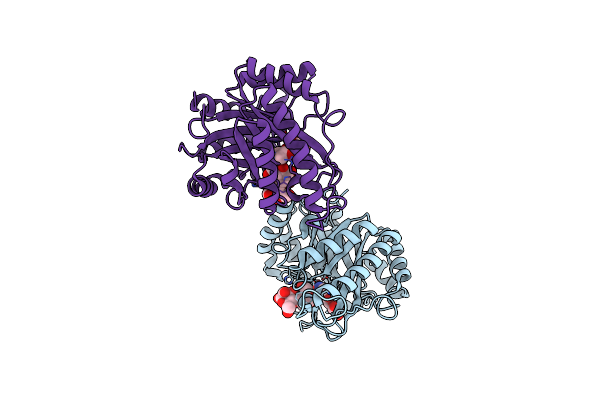 |
The Structure Of Gh113 Beta-Mannanase Aamana From Alicyclobacillus Acidocaldarius In Complex With Manifg And Beta-1,4-Mannobiose
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-04-02 Classification: HYDROLASE Ligands: IFM, BMA |
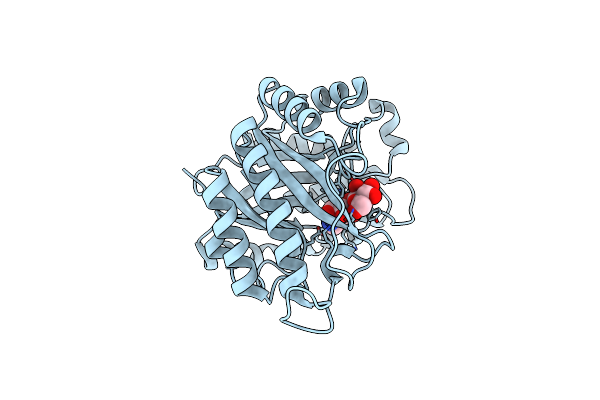 |
The Structure Of Gh113 Beta-Mannanase Aamana From Alicyclobacillus Acidocaldarius In Complex With Manmim
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2014-04-02 Classification: HYDROLASE Ligands: MVL, BMA |
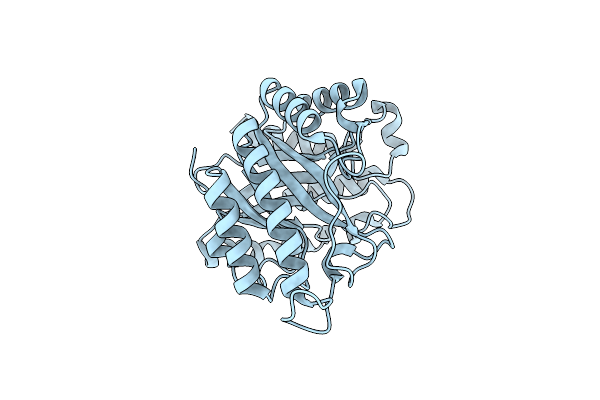 |
Crystal Structure Of The Endo-Beta-1,4-Mannanase From Alicyclobacillus Acidocaldarius
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-08-26 Classification: HYDROLASE |
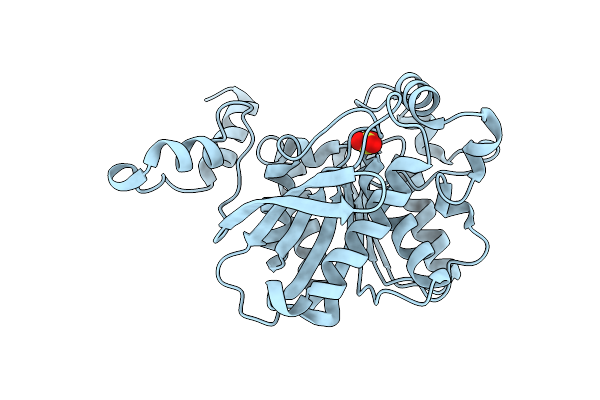 |
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-06-26 Classification: HYDROLASE Ligands: SO4 |
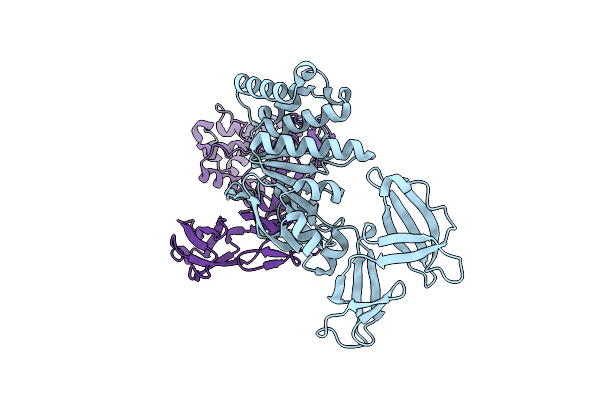 |
Structure Of The Atpase Subunit Cysa Of The Putative Sulfate Atp-Binding Cassette (Abc) Transporter From Alicyclobacillus Acidocaldarius
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2005-06-07 Classification: LIGAND BINDING PROTEIN Ligands: CL |
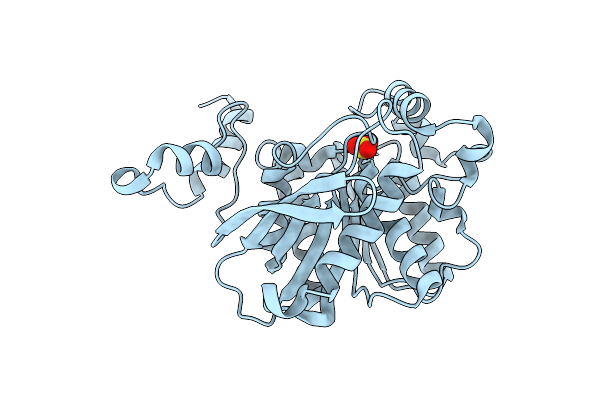 |
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-10-05 Classification: HYDROLASE Ligands: SO4 |
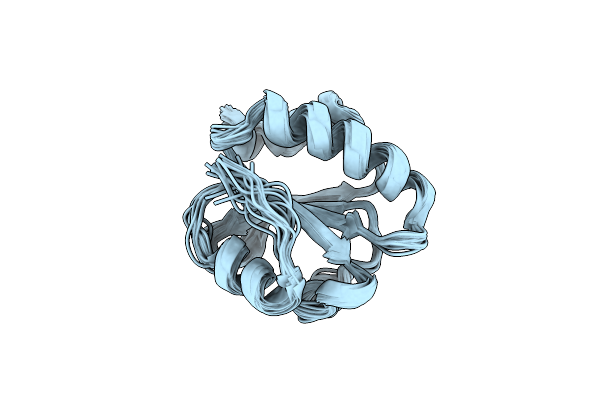 |
Solution Structure Of The K18G/R82E Alicyclobacillus Acidocaldarius Thioredoxin Mutant
Organism: Alicyclobacillus acidocaldarius
Method: SOLUTION NMR Release Date: 2004-06-22 Classification: OXIDOREDUCTASE |
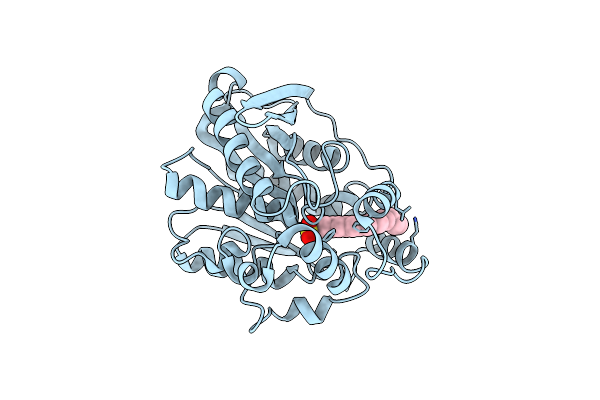 |
Crystal Structure Of Mutant M211S/R215L Of Carboxylesterase Est2 Complexed With Hexadecanesulfonate
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-03-23 Classification: HYDROLASE Ligands: HDS |
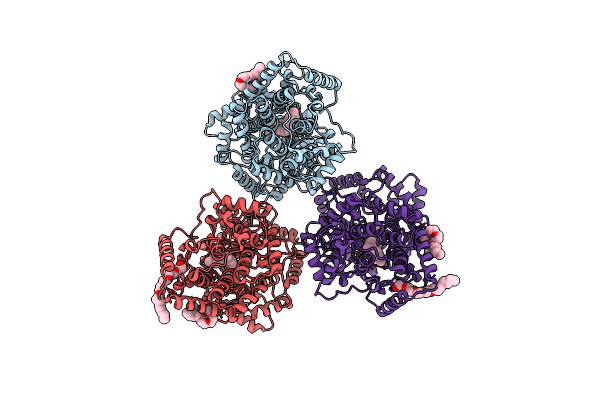 |
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2004-02-03 Classification: ISOMERASE Ligands: C8E, SQA |
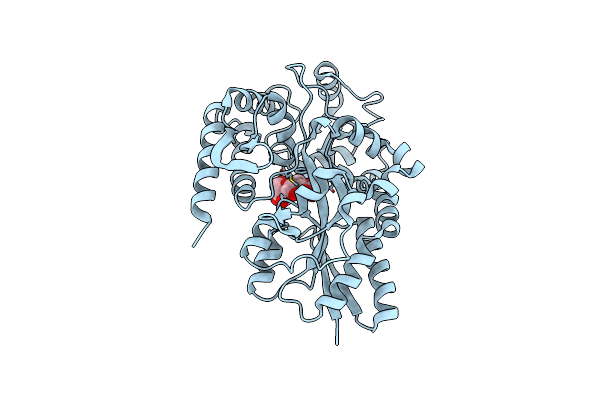 |
X-Ray Structures From The Maltose-Maltodextrin Binding Protein Of The Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2003-12-11 Classification: MALTOSE-BINDING PROTEIN |
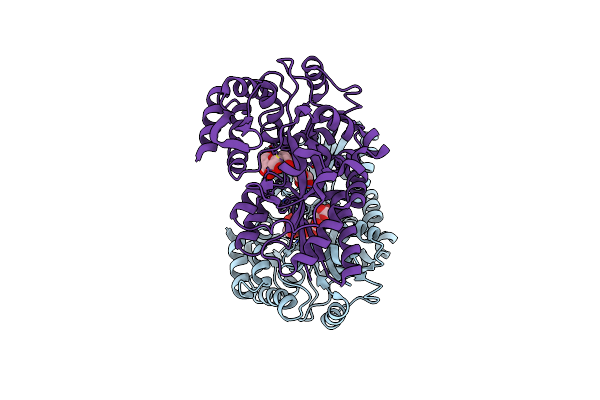 |
X-Ray Structures Of The Maltose-Maltodextrin Binding Protein Of The Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2003-12-11 Classification: MALTOSE-BINDING PROTEIN |
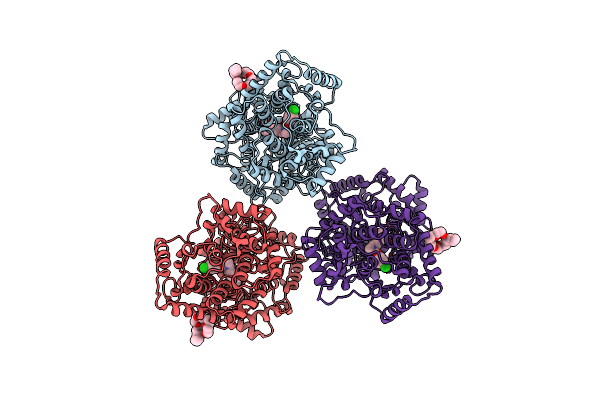 |
Structures Of Human Oxidosqualene Cyclase Inhibitors Bound To An Homologous Enzyme
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2003-10-23 Classification: ISOMERASE Ligands: C8E, R23 |
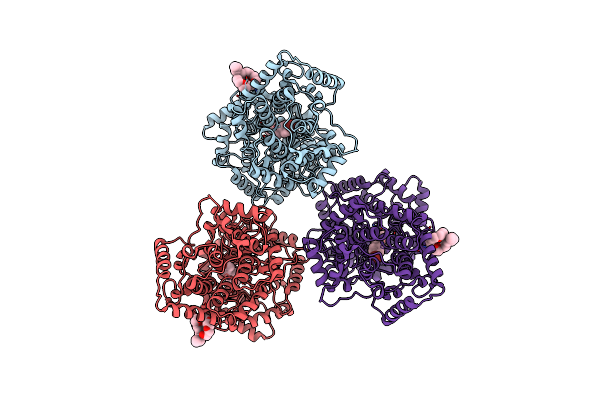 |
Structures Of Human Oxidosqualene Cyclase Inhibitors Bound To An Homologous Enzyme
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2003-10-17 Classification: ISOMERASE Ligands: C8E, R17 |
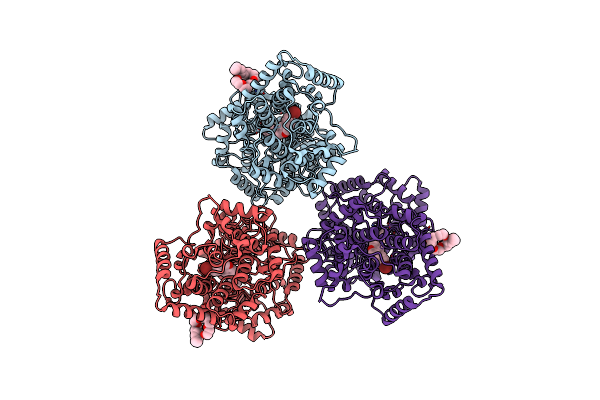 |
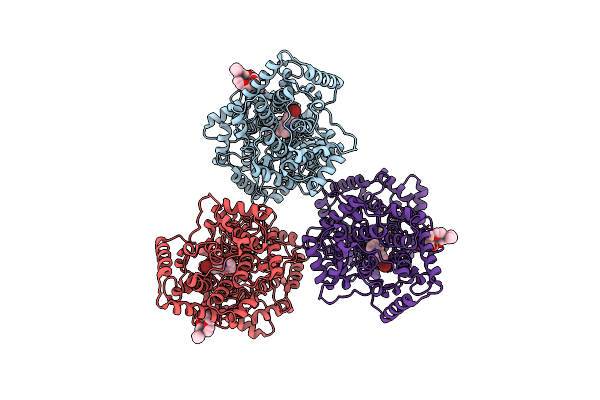
Structures Of Human Oxidosqualene Cyclase Inhibitors Bound To An Homologous Enzyme
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2003-10-17 Classification: ISOMERASE Ligands: C8E, R19 |
 |
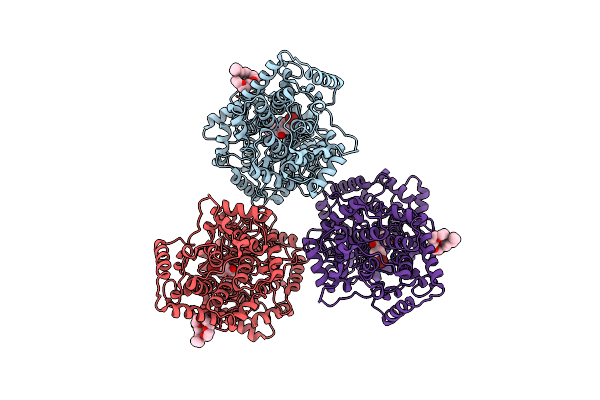
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2003-10-02 Classification: ISOMERASE Ligands: C8E, W37 |
 |
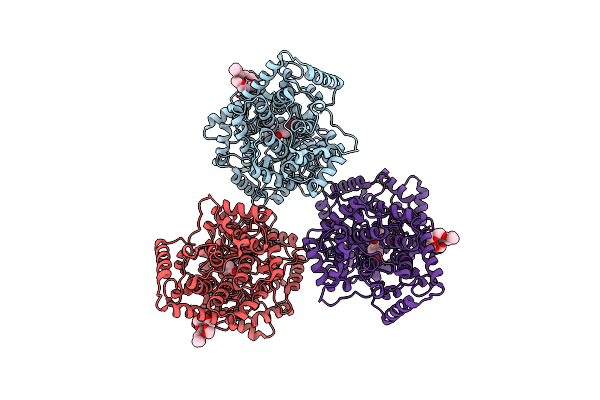
Structures Of Human Oxidosqualene Cyclase Inhibitors Bound To An Homologous Enzyme
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2003-08-21 Classification: ISOMERASE Ligands: C8E, R01 |
 |
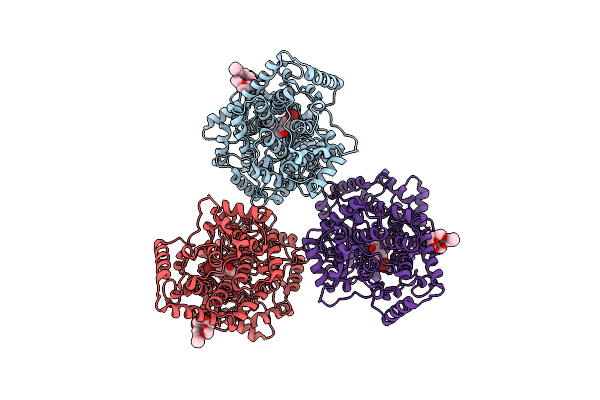
Structures Of Human Oxidosqualene Cyclase Inhibitors Bound To An Homologous Enzyme
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2003-08-21 Classification: ISOMERASE Ligands: C8E, R88 |
 |
Structures Of Human Oxidosqualene Cyclase Inhibitors Bound To An Homologous Enzyme
Organism: Alicyclobacillus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2003-08-21 Classification: ISOMERASE Ligands: C8E, R02 |