Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
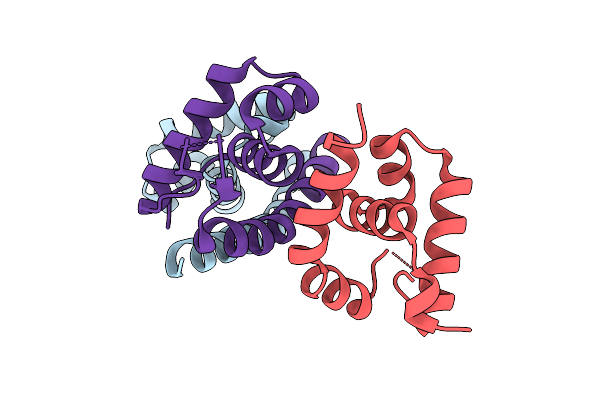 |
Crystal Structure Of The Acinetobacter Baumannii Lysr Family Regulator Acer Dna-Binding Domain (P321)
Organism: Acinetobacter baumannii (strain atcc 17978 / dsm 105126 / cip 53.77 / lmg 1025 / ncdc kc755 / 5377)
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: DNA BINDING PROTEIN |
 |
Organism: Legionella pneumophila str. corby
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: HYDROLASE Ligands: A1IRK, ZN |
 |
Organism: Legionella pneumophila str. corby
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: HYDROLASE Ligands: A1IRL, BU3, GOL, ZN, SO4 |
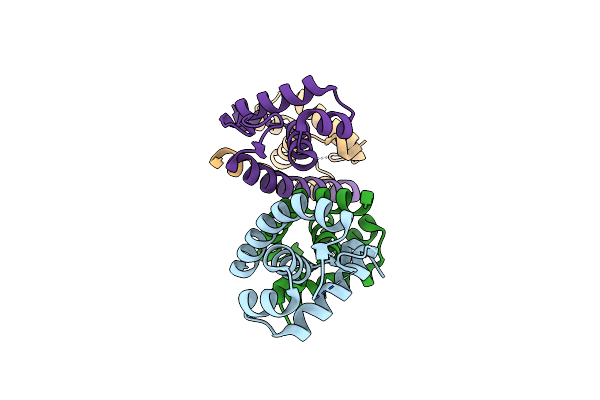 |
Crystal Structure Of The Acinetobacter Baumannii Lysr Family Regulator Acer Dna-Binding Domain (H32)
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2025-04-16 Classification: TRANSCRIPTION |
 |
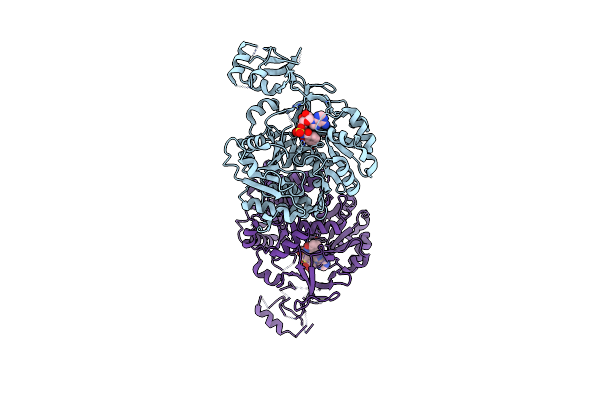
Structure Of Fbsh, An Nrps Adenylation Domain In The Fimsbactin Biosynthetic Pathway Bound To 2,3-Dihydroxybenzoic Acid.
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-11-20 Classification: LIGASE/SUBSTRATE Ligands: DBH, EDO |
 |
Structure Of Fbsh, An Nrps Adenylation Domain In The Fimsbactin Biosynthetic Pathway Bound To Salicyl-Ams
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2024-11-20 Classification: LIGASE/INHIBITOR Ligands: KT0, EDO |
 |
Organism: Acinetobacter baumannii atcc 17978
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-11-29 Classification: TRANSFERASE Ligands: GOL, NA |
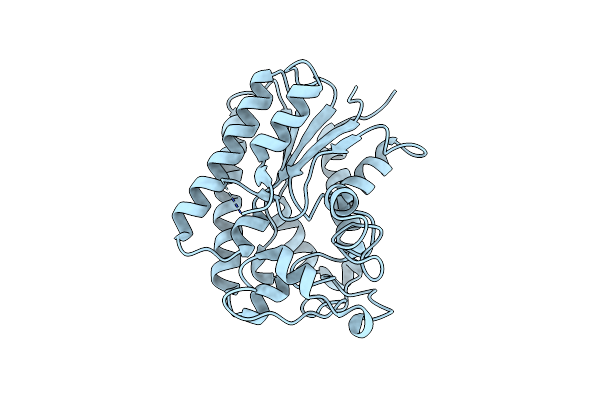 |
Lysophospholipase Plaa From Legionella Pneumophila Str. Corby - Iodide Soak
Organism: Legionella pneumophila str. corby
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2023-06-14 Classification: HYDROLASE Ligands: IOD |
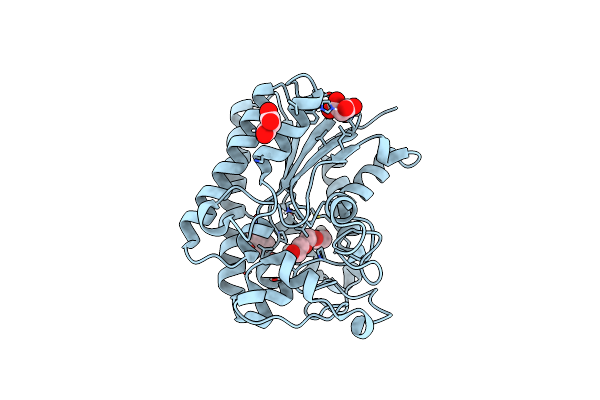 |
Lysophospholipase Plaa From Legionella Pneumophila Str. Corby - Complex With Peg Fragment
Organism: Legionella pneumophila str. corby
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2023-06-14 Classification: HYDROLASE Ligands: MLA, BU3, PGE, IOD |
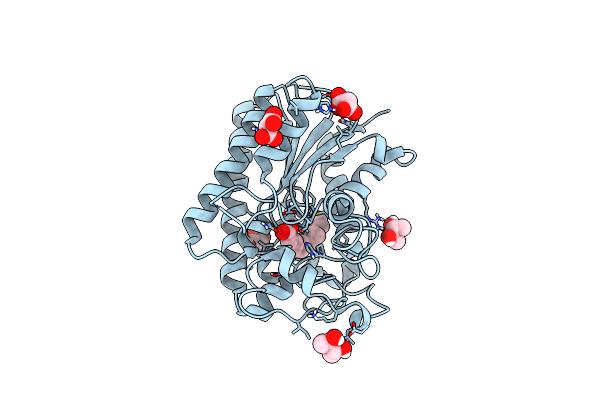 |
Lysophospholipase Plaa From Legionella Pneumophila Str. Corby - Complex With Palmitate
Organism: Legionella pneumophila str. corby
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-06-14 Classification: HYDROLASE Ligands: MLA, PLM, BU3 |
 |
Structure Of 6-Carboxy-5,6,7,8-Tetrahydropterin Synthase Paralog Qued2 From Acinetobacter Baumannii
Organism: Acinetobacter baumannii atcc 17978
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-12-07 Classification: LYASE Ligands: EDO, ZN, PEG |
 |
Crystal Structure Of The Dsrbd Domain Of Trna-Dihydrouridine(20) Synthase From Amphimedon Queenslandica
Organism: Amphimedon queenslandica
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2022-11-30 Classification: RNA BINDING PROTEIN Ligands: BME, CL, GOL |
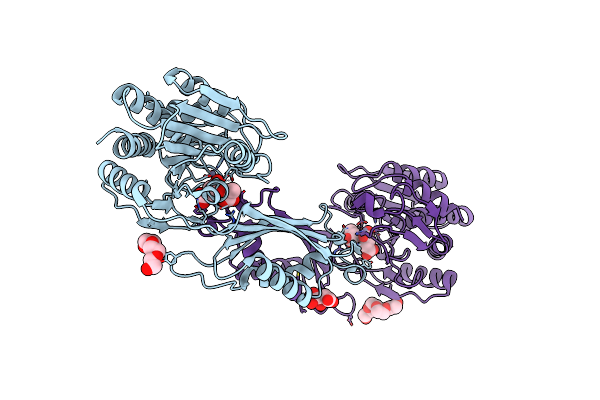 |
Crystal Structure Of The Succinyl-Diaminopimelate Desuccinylase (Dape) From Acinetobacter Baumannii In Complex With Succinic And L-Lactic Acids
Organism: Acinetobacter baumannii atcc 17978
Method: X-RAY DIFFRACTION Release Date: 2022-11-30 Classification: HYDROLASE Ligands: ZN, SIN, 2OP, PGE, PG4, CIT |
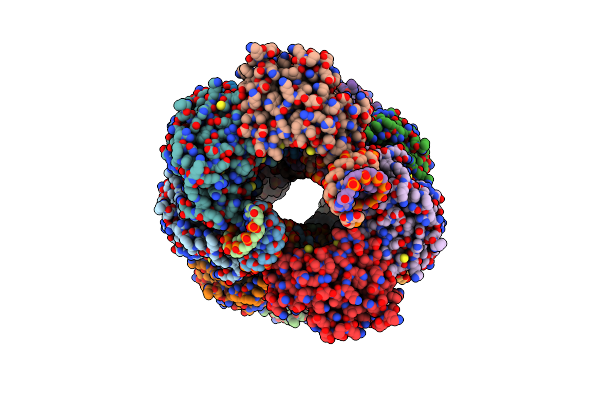 |
Organism: Dna molecule, Linum usitatissimum
Method: ELECTRON MICROSCOPY Release Date: 2022-06-08 Classification: HYDROLASE/DNA |
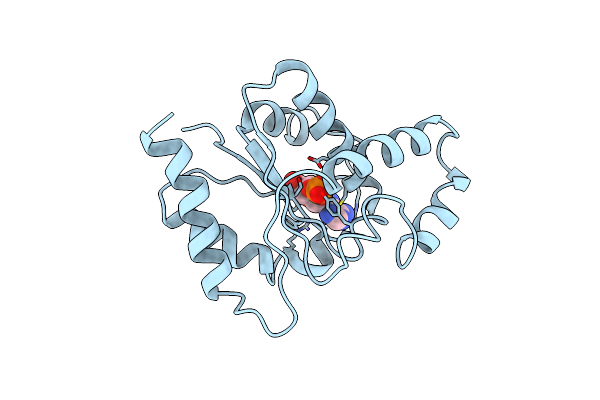 |
Organism: Linum usitatissimum
Method: ELECTRON MICROSCOPY Release Date: 2022-06-01 Classification: HYDROLASE Ligands: ACK |
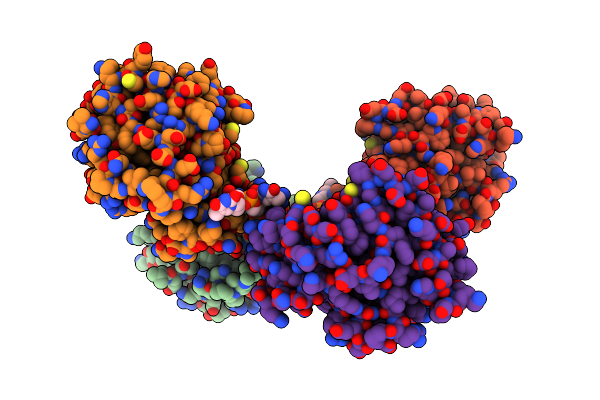 |
Organism: Dna molecule, Linum usitatissimum
Method: ELECTRON MICROSCOPY Release Date: 2022-06-01 Classification: HYDROLASE/DNA |
 |
Organism: Dna molecule, Linum usitatissimum
Method: ELECTRON MICROSCOPY Release Date: 2022-06-01 Classification: HYDROLASE/DNA Ligands: ACK |
 |
Organism: Linum usitatissimum
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2022-02-09 Classification: LYASE Ligands: NAD, ZN, GOL, MG, EDO, BTB |
 |
Crystal Structure Of Hydroxynitrile Lyase From Linum Usitatissimum Complexed With Acetone Cyanohydrin
Organism: Linum usitatissimum
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2022-02-09 Classification: LYASE Ligands: NAD, ZN, CNH, MG, EDO, GOL, BTB, ACN |
 |
Crystal Structure Of Hydroxynitrile Lyase From Linum Usitatissium Complexed With (R)-2-Hydroxy-2-Methylbutanenitrile
Organism: Linum usitatissimum
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2022-02-09 Classification: LYASE Ligands: NAD, ZN, MG, 5Z9, EDO, GOL, BTB |