Search Count: 16
 |
Crystal Structure Of Holo-Swhpa-Mg (Hydroxy Ketone Aldolase) From Sphingomonas Wittichii Rw1 In Complex With Hydroxypyruvate And D-Glyceraldehyde
Organism: Rhizorhabdus wittichii rw1
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-11-23 Classification: LYASE Ligands: 3PY, 3GR, PEG, MG, BR, K |
 |
Crystal Structure Of S116A Mutant Of Hydroxy Ketone Aldolase (Swhka) From Sphingomonas Wittichii Rw1 In Complex With Hydroxypyruvate
Organism: Sphingomonas wittichii (strain rw1 / dsm 6014 / jcm 10273)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-11-16 Classification: LYASE Ligands: 3PY, MG |
 |
Crystal Structure Of Holo-H44A Mutant Of Hydroxy Ketone Aldolase (Swhka) From Sphingomonas Wittichii Rw1, In Complex With Hydroxypyruvate
Organism: Sphingomonas wittichii (strain rw1 / dsm 6014 / jcm 10273)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-11-16 Classification: LYASE Ligands: MG, BR, K, 3PY |
 |
Crystal Structure Of Holo-F210W Mutant Of Hydroxy Ketone Aldolase (Swhka) From Sphingomonas Wittichii Rw1 In Complex With Hydroxypyruvate
Organism: Sphingomonas wittichii (strain rw1 / dsm 6014 / jcm 10273)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-10-26 Classification: LYASE Ligands: 3PY, PEG, MG, K, BR |
 |
Crystal Structure Of Holo-F210W Mutant Of Hydroxy Ketone Aldolase (Swhka) From Sphingomonas Wittichii Rw1, With The Active Site In The Resting And The Active State
Organism: Sphingomonas wittichii (strain rw1 / dsm 6014 / jcm 10273)
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-10-26 Classification: LYASE Ligands: MG, K, 3PY |
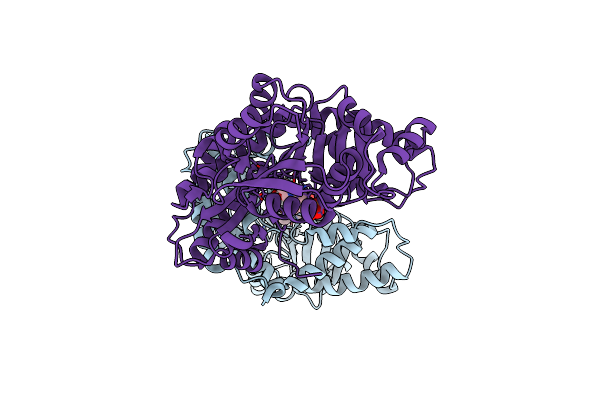 |
Alanine-Glyoxylate Aminotransferase 1 (Agt1) From Arabidopsis Thaliana In Presence Of Serine
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-10-23 Classification: TRANSFERASE Ligands: PLP, 3PY |
 |
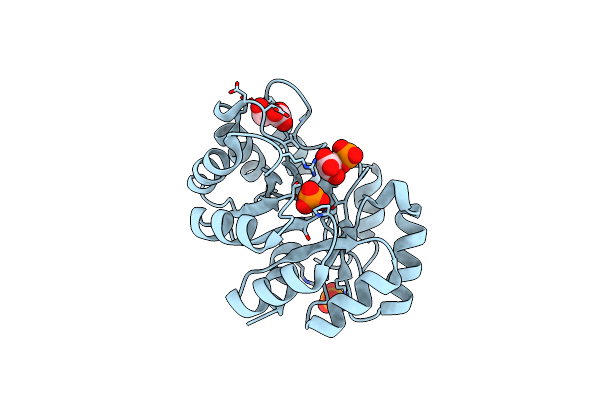
Crystal Structure Of A Class Ii Pyruvate Aldolase From Sphingomonas Wittichii Rw1 In Complex With Hydroxypyruvate
Organism: Sphingomonas wittichii rw1
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-05-29 Classification: LYASE Ligands: 3PY, ACT, MG, BR, K, NA |
 |
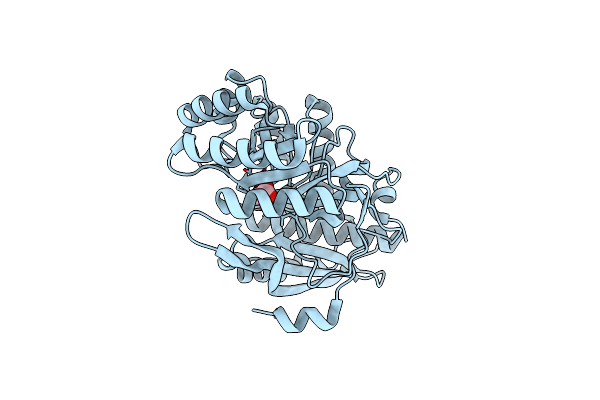
Crystal Structure Of The 2-Keto-3-Deoxy-6-Phosphogluconate Aldolase Of Zymomonas Mobilis Zm4 With 3-Phosphoglycerate
Organism: Zymomonas mobilis subsp. mobilis (strain atcc 31821 / zm4 / cp4)
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2018-06-20 Classification: LYASE Ligands: PO4, 3PG, 3PY |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2015-10-14 Classification: ISOMERASE Ligands: 3PY, MG |
 |
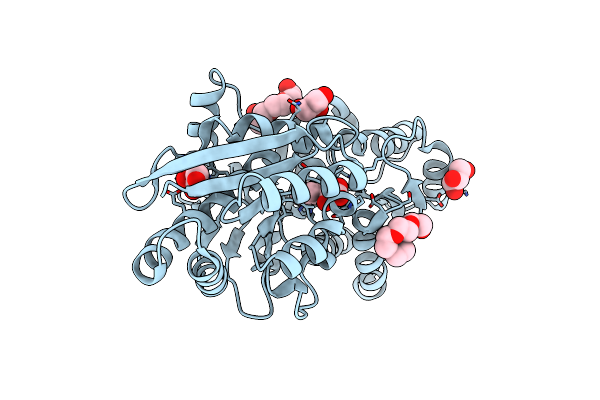
Structure Of The Carboxyl Transferase Domain From Rhizobium Etli Pyruvate Carboxylase With 3-Hydroxypyruvate
Organism: Rhizobium etli
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2013-11-13 Classification: LIGASE Ligands: ZN, 3PY, BTN, MG, CL, GOL |
 |
The Crystal Structure Of A Transporter In Complex With 3-Phenylpyruvic Acid
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-02-29 Classification: SIGNALING PROTEIN Ligands: PPY, GOL, 3PY, PG4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2011-01-19 Classification: LYASE Ligands: 3PY, SO4 |
 |
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2004-01-13 Classification: ISOMERASE Ligands: PO3, 3PY, 2PG |
 |
Crystal Structure Of N-Acetylneuraminate Lyase From E. Coli Mutant L142R In Complex With B-Hydroxypyruvate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2003-05-09 Classification: LYASE Ligands: 3PY |
 |
X-Ray Structure Of Desulfovibrio Desulfuricans Bacterioferritin: The Diiron Centre In Different Catalytic States (As-Isolated Structure)
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2003-04-01 Classification: IRON STORAGE/ELECTRON TRANSPORT Ligands: FE, SO4, GOL, FEC, 3PY |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 1997-10-22 Classification: LYASE Ligands: 3PY |

