Search Count: 7
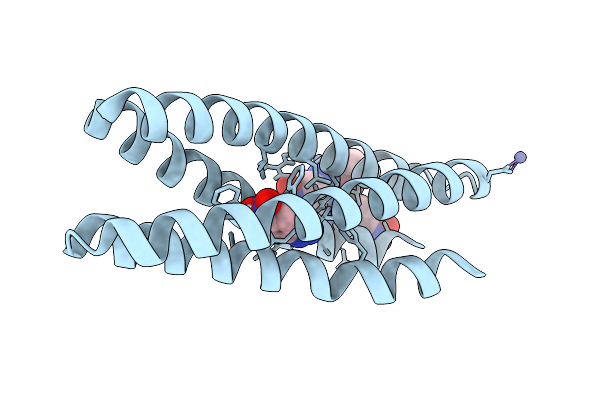 |
Crystal Structure Of Kable Double Mutant (Lys14Pro, Leu20Trp) In Complex With 6-Nitrobenzotriazole
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: DE NOVO PROTEIN Ligands: 3NY, CAC, ZN |
 |
Crystal Structure Of Kemp Eliminase 1A53-Core With Bound Transition State Analogue
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-01-10 Classification: LYASE Ligands: PO4, 3NY |
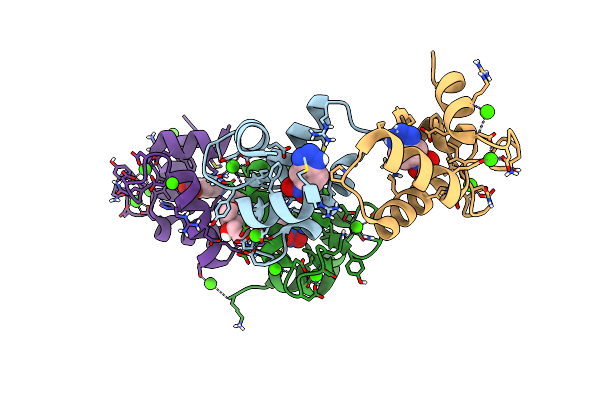 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-07-27 Classification: BIOSYNTHETIC PROTEIN/INHIBITOR Ligands: 3NY, CA, MPD |
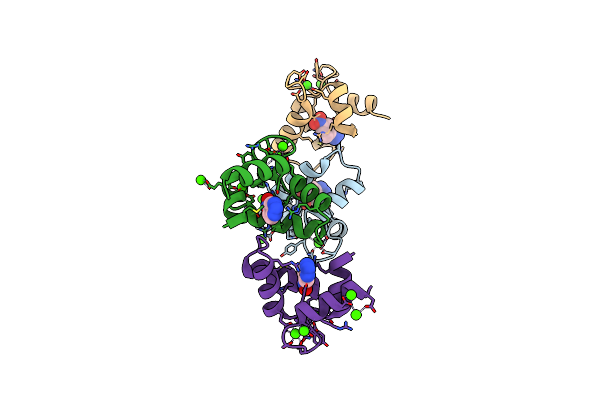 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-07-27 Classification: BIOSYNTHETIC PROTEIN/INHIBITOR Ligands: 3NY, CA |
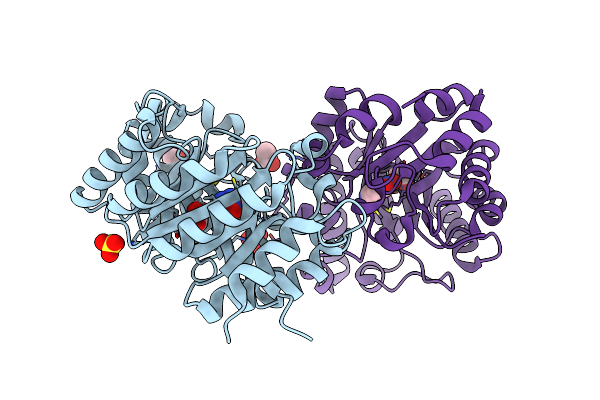 |
Crystal Structure Of Kemp Eliminase Hg-2 Complexed With Transition State Analog 5-Nitro Benzotriazole
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2011-06-29 Classification: HYDROLASE Ligands: 3NY, ACT, SO4 |
 |
Crystal Structure Of Kemp Elimination Catalyst 1A53-2 Complexed With Transition State Analog 5-Nitro Benzotriazole
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2011-06-29 Classification: LYASE Ligands: 3NY, SO4, TLA |
 |
Optimization Of The In Silico Designed Kemp Eliminase Ke70 By Computational Design And Directed Evolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2011-02-09 Classification: LYASE Ligands: 3NY |

